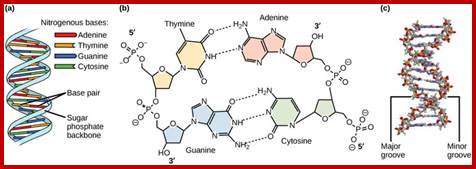
Structure of DNA:
In order to understand the structure and dynamics of DNA, it is good to begin with the properties of Nucleic acid and nucleotides; and how nucleotides are linked to each other and the conformations of sugars; finally, how the nucleotides base pair. Formation of polynucleotide chains in sequence specific mode requires another polynucleotide chain as a template. When life originated around ~3.8 billion years ago, in the then chemical soup nucleotides developed and then they are randomly joined into ribo-polynucleotide chains. Millions of such configurations in our pre-biological world are generated in the fresh oceanic water, (where and when) among them some are naturally chosen and selected which could perform catalysis and replication. This is chemical evolution. As RNAs were formed first and found to be unstable and unable to build-in properly for replication as they also have enzyme properties they copied their genomic RNA chains into DNA in 5’ to 3’, and copied DNA was found to be more stable. Many of such genomic DNAs have survived and further changed/evolved by acquiring changes and developed new templates with different Nucleotide sequences, during the course of billion years. As single stranded DNA genomes were unstable they developed two stranded DNA complementary to each other; it is an enigma. Is there any role of RNA as an enzyme? But RNAs used their template to assemble amino acids to generate polypeptides. In spite of RNA gave place to DNA as the genomic material, without RNA, DNA cannot replicate itself without the help of another important functional molecule called protein. Generation of proteins is found to be an important step in chemical evolution. All the three are polymers of nucleotides (DNA and RNA) and amino acids (proteins). More than 99% of organisms including viruses possess DNA as their genome, yet some viruses contain single stranded or double stranded RNA as their genomes.
RNA and DNA genomes use the template and their sequence to generate a complimentary copy of the same. During nucleotide assembly, covalent phosphodiester bonds form each time they add by one nucleotide and the chain always grew in 5’ to 3’ direction. Whether it is single stranded DNA or double stranded DNA, for their synthesis they require templates and specific enzymes. One of the enzyme called polynucleotide phosphorylase is capable of polymerization of tri-phospho nucleotides to polynucleotides chains randomly, so also they can phosphorolyse them; it can also degrade the RNA chain.
Polymers of DNA exists in single stranded form, double stranded form and occasionally they are also found in three stranded or four stranded structures. The structure can assume various forms depending upon the sequence; it is this feature that ultimately determines the DNA structural form. They can exist either in linear form or circular form, the structure can be bent or curved, or flexible, or its helical structure can be can be left handed or right handed coiled; thus, a DNA in cells don’t exhibit only one form, but it exists in different structural (segments) forms throughout the length of the DNA.

Spilled out Viral DNA; DNA spilled out of bacterial;
Gopal Murti; http://www.allposters.com/
The First, dsDNA Structure was Proposed by J. D. Watson and F. H. Crick (April, 1953):
|
|
|
Genetical
Implications of the structure of Deoxyribonucleic Acid |
|
Six papers were published by “The Great Nature” in the same volume and the same year on the structure of DNA-by- Watson J.D and Crick F.H.C: Wilkin and Miss Franklin, Williams M.H.F; Stokes A.R. and Wilson H.R et al; Franklin R and Gosling R.J, Two papers by Watson and crick- below-
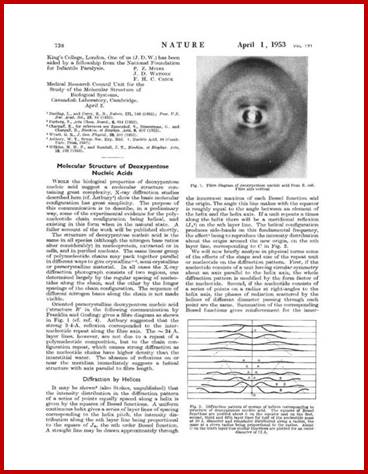
Watson and Crick’ corrected certain mistakes in the paper (mistakes regretted); (Watson, J. D., and Crick, F. H. C., 1953. Nature 171:) Nature has now re-issued a corrected version of the paper, copies of which you can find in the above figure. First paper;

Watson and Crick’s famous revised paper, in its entirety. Second paper; (Watson, J. D., and Crick, F. H. C., 1953. Nature 171:737-738.)
SsDNA:
· Many viruses have single stranded DNAs as their genome; they can be either circular or linear ex. PhiX174 or M13 single stranded, circular. Linear ssDNA of Parvoviridae (-) sense, HIV unintegrated RNA (+) sense. All single stranded linear DNA and RNA molecules have 5’ to 3’ polarity. Single stranded linear DNA viruses are Parvovirus like; ex. Bombyx mori densovirus type 2, Hepatopancreatic parvo-like virus and Lymphoidal parvo-like virus.
· ssRNA can exist either in positive or negative sense; (positive DNA Picorna virus circular; negative sense DNA Mononegavirales include nonsegmented (-) strand;
· When suspended in aqueous solutions they show random coiled features, for each of the nucleotides have bases linked to their sugars by glycosidic bonds at C-1’ position in such a way their sugars show C2 endo and C3 exo puckering, hence they show anti configuration.
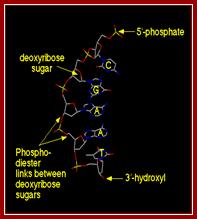
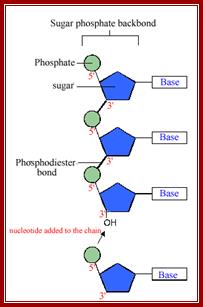
· Sugar phosphate is the backbone, where Phosphate group provides negative charges and H2O binds to Phosphate groups and they push sugar-base inwards of the linear polynucleotide chain.
· In addition, as hydrophobic bases being flat, rigid structures, they project out at right angles to phosphate–sugar–phosphate backbone. The C2’ endo and C3’exo sugar puckering with anti-configuration of phosphate group and base with respect to sugar puckering, renders ssDNA with right handed helicity, which always changes every moment in water. Understanding sugar puckering is important in understanding the DNA structure. Hydrophobic interactions between complementary bases lead to base pairing of flat bases by hydrogen bonds. The flat bases one above the other produces torsional twist.
Comparison of DNA and RNA Nucleotides-sugar phosphates with sugar puckering;

3’endo and 2’exo in N-type and 2’endo and 3’exo in DNA; http://www.biosyn.com
Single stranded DNA; http://www.bic.arizona.edu
Single stranded DNA; http://click4biology.info/
· Interestingly phosphate in PSP strand pulled outside by H2O and hydrophobic bases pulled inwards for their hydrophobic forces. They projected at right angles to P-S-P backbone, as the bases have hydrophobic features, the bases get attracted to each other by hydrophobicity thus the PSP strand is pulled inwards, so the back-bone bends and coils by forces such as propeller twist. Phosphate group at periphery has negative charges that attracts water, so it pulls the PSP backbone outwards. Base pairs pull the PSP inwards. So, the ds DNA strands take twist in right handed direction. Inside the coil the base pairs being hydrophobic no water enters the central core of the helix.
· SsDNA strands also exhibit duplex or hair pin or stem loop structural features wherever the sequences are complementary to each other, such secondary structures have rotational constraints, at the same time non-Watson-Crick’s interaction can lead to folding of DNA into a compact 3-D structure as in protein (not observed so far?).
· Most of ssDNA genomes, either linear or circular, are generally packed into viral proteinaceous coats or capsids.
· In host cells, they exhibit super coiled states, associated proteins provide stability.
· The genomic ssDNAs show either positive or negative sense characters. Positive sense is that their sequence is complementary to that of mRNA. The negative sense is that the sequence is same as that of mRNAs.
· Replication of ssDNA is through ds-replicative form.
Insights into ds DNA structures:
The structural feature of ds DNA posed perplexing problems and paradoxes to the pioneers of structural biologists. Paradoxically it is not the structural chemists who solved the 3-D structure, but it is the biologists responsible for solving the most extraordinary and fascinating structure of DNA, the mother of all life forms, with certain exceptions.
Scientists from various labs from different continents worked towards understanding of the DNA structure. People working in Cavendish lab and King’s College, London and scientists in USA have immensely contributed in elucidating the basic structural features. Notable among them are F. Crick, J. D. Watson, and M. Wilkins, R. Franklin, Linus Pauling and Williams M.H.F, Gosling and Stokes A.R few others. Though the available information on DNA was made known to others, some, for strategic purposes the core information held back by R. Franklin and M. Wilkins and Gosling. Each bit of information that was made available was like scrambled bits, every body knew some aspects of the structure, but none were able put them together the jigsaw puzzle to its entirety and the quintessence of it remained a secret. Even L. Pauling could not guess the correct structural features in spite of being a good biochemical scientist, but published papers on the same subject in “Nature”.
Maurice Wilkins and Miss Roslyn Franklin in 1950s had the best possible X-ray diffraction data of liquid DNA threads. Unfortunately, they were unable to discerningly understand, and put the facts into a structural model. The drawback for them was, they being biophysicists, (crystallographers) never thought like biologists; for biologists never think like a crystallographer, but inheritance was the key for them, and the key was duplication of the genetic material at the time of birth, growth and reproduction. For Biophysist the crystalline structure and crystals were important. Today all fields of science have to contribute to understand anything of science.
Francis Harry Compton Crick, brilliant biophysicists, a war veteran, and an erudite on X-ray diffraction studies, he was like a grandpa to the young and bubbling innovator and a motivator of persons like James D. Watson.
Dr. J.D. Watson was a Ph.D. (Phage Genetics) under the guidance of great Luria (X-ray effect on bacteriophage) from USA. During one of the symposiums he met Maurice Wilkins, it is then he saw the X-ray diffraction pictures of DNA (taken by Miss Franklin). Then he moved to Cavendish lab in 1951. He was doing his postdoctoral studies in England; he was a biologist par excellent and a romantic too.
He did not understand anything (?) when he first saw X-ray diffraction pictures; as any other biologist (like me) could make out or discern; he saw a black and white photograph with black dots and patches in a set pattern that was all the first impression for him (Like me, a stupid). He was not new to X-ray diffraction though. What prompted Dr. Watson to further the studies on DNA structures is any body’s guess. His interaction with Crick, Wilkins, and many others involved in DNA studies was infectious. Watson gained some important insights into the structural chemistry of DNA, which is important. Perhaps he worked on the simple basic biochemistry to understand the structures of phosphates, sugars, bases and their interaction and chemical bond formation. Understanding of basic biochemistry is very important for any molecular biologist or for any biologists. His book called Molecular Biology of the Gene explains the biochemistry of DNA. The most puzzling aspect dsDNA was which base paired with which base. The answer for this was available for Chargaff has published his paper on the chemistry of Nucleic acid, where he found the quantity of Adenine found equal to Thymidine and Guanine equal to cytosine.

Levene’s model; Levene was correct in identifying the three parts of a nucleotide, and determining that nucleotides were linked together to make DNA; however, in 1928, he also incorrectly proposed that one of each of the four nucleotides was linked together in a small circular molecule and that these "tetra nucleotides" were the basis of DNA (Levene and London,1928)
Every time the story of DNA structure seems to reach a conclusion, it bounces back to center stage yet another incarnation developed. The latest avatar to manifest itself is a stretched and overwound form of DNA reported recently by a French group1, working with single DNA molecules. When a moderately large stretching force (of about 3 Pico Newtons) is applied, the DNA molecule apparently becomes highly twisted and extended, but even more amazingly it takes up an ‘inside-out structure’ in which the phosphodiester chain is on the inside and the bases are exposed.
Precisely such a structure was proposed in 1953 by Pauling and Corey2 (albeit with three strands), making use of ‘the general principles of molecular structure’, which they had applied with such spectacular success in predicting the a -helix and b -structure for proteins. While they were convinced that the DNA structure was a helix, they did not make use of the already available chemical data of Chargaff, which demonstrated that although the base composition of DNA varies widely, certain bases were always present in equal numbers (viz. Adenine = Thymine and Guanine = Cytosine), clearly suggesting some kind of pairing between the bases; DNA structure another Avatar; http://www.iisc.ernet.in; www.pinsdaddy.com

A premature hypothesis by Watson and Crick of the structure of DNA base pairing. The phosphate groups are pointing inwards binding to central Mg2+ and the bases outwards. (Adopted from: The Double Helix, by J. Watson); even Pauling did that, bases projecting out of the phosphate-sugar backbone and phosphate groups join each other; http://www.forensicgenealogy.info

Linus Pauling speaking in a lecture hall with his protein Model;
https://profiles.nlm.nih.gov

Triple helix-DNA
model by Linus Pauling; Although the chemistry was wrong, Linus
Pauling's triple-stranded DNA model was a catalyst for James Watson and Francis
Crick to solve the structure of DNA.
Originally created for DNA Interactive; ( http://www.dnai.org ), Triple
stranded DNA model; Pauling and Corey; PNAS-1953 Feb; http://scarc.library.oregonstate.edu;
http://www.gettyimages.com
L .Pauling- Nobel prize in 1954 (chemistry) and 1962 (peace).Author of books (landmark book The nature of Chemical bonds, and 1000 articles; http://lpi.oregonstate.edu

Pauling and Corey; sugar phosphates are in the center of the coil and nucleotide bases are projecting out; http://scarc.library.oregonstate.edu; http://www.dnacoil.com

Linus Pauling's triple stranded Helix DNA model where three strands intertwined, https://www.dnalc.org

L. Pauling with his wife and children; http://www.gettyimages.co.uk

Open-relaxed and supercoiled DNA molecules; https://www.biochem.wisc.edu; http://www.cbs.dtu.dk/

The most celebrated X-ray diffraction photograph of liquid crystal (LCD) of DNA taken by Miss Franklin’ and Raymond Gosling (1950s) which provided the food for thought and gave all the insights for others to get the knowledge of dsDNA structure and Nobel Laureate honors, for she never thought of understanding and the importance DNA structure and she missed it but not her work, for she was in heaven; Nika Knight. http://www.corriere.it.

Rosalind Elsie Franklin- ‘The Queen of Molecular Biology’; X-ray diffraction of DNA crystals. https://www.livescience.com; https://www.pinterest.com
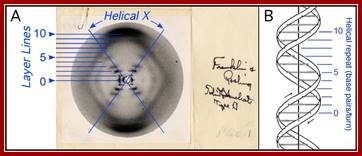
A single crystal structure-‘photo 51’ Franklin’s; Structure of B-DNA. A. Photograph 51 of B-DNA. X-ray diffraction photograph of a DNA fiber at high humidity taken by Franklin and Gosling, 1953b. Interpretation of the helical-X and layer lines added in blue. B. Watson- Crick model of B-DNA, adopted from (Watson and Crick, 1953b), with the helical repeat associated with the layer lines labeled. https://www.reddit.com
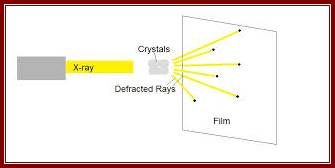
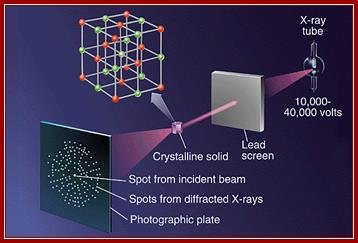
Constructive interference indicates that the diffracted x-rays are in phase or lined up with each other, while destructive interference indicates that the x-rays are not exactly in phase with each other. The result is that the measured intensity of the x-rays increases and decreases as a function of angle and distance between the detector and the crystal. www.en.wikibooks.org;
The x-rays that have been scattered in various directions are then caught on x-ray film, which show a blackening of the emulsion in proportion to the intensity of the scattered x-rays hitting the film, or by a solid-state detector, like those found in digital cameras. The crystal is rotated so that the x-rays are able to hit the protein from all sides and angles. The pattern on the emulsion reveals much information about the structure of the protein in question. The three basic physical principles underlying this technique are:
1. Atoms scatter x-rays. The amplitude of the diffracted x-ray is directly proportional to the number of electrons in the atom.
2. Scattered waves recombine. The beams reinforce one another at the film if they are in phase or cancel one another out if they are out of phase. Every atom contributes to a scattered beam.
3. Three-dimensional atomic arrangement determines how the beams recombine.
The intensities of the spots and their positions are thus the basic experimental data of the analysis. The final step involves creating an electron density map based on the measured intensities of the diffraction pattern on the film. A Fourier Transform can be applied to the intensities on the film to reconstruct the electron density distribution of the crystal.
How to use X-ray diffration and get the result.
The position of the spots observed in X-ray diffraction are determined by Braggs Law: n lambda =2D sin (Φ);Crystals; www.Learneasy.info
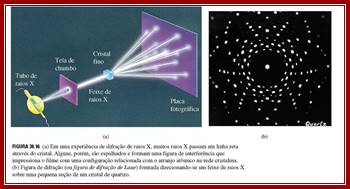

Left, the major steps involved in DNA structure determination by X-ray crystallography showing the important role played by molecular models of DNA structure in this iterative process. Right, an image of actual A- and B- DNA X-ray patterns obtained from oriented and hydrated DNA fibers (courtesy of Dr. Herbert R. Wilson, FRS- see refs. list).

Nobel Foundation; (C) Crick, Watson, & Wilkins; http://jeromeprophet.blogspot.in/

From
left, Rosalind Franklin in
1956, James Watson in
the 1980s, Francis Crick
in the 1980s, and Maurice Wilkins in the early
1990s. Franklin died in 1958.
Both Crick and Wilkins died in 2004. ; http://www.forensicgenealogy.info/

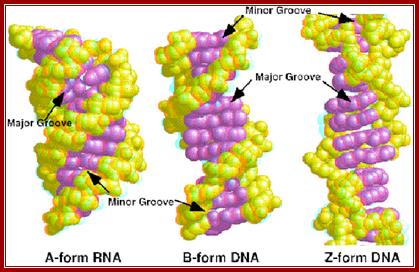
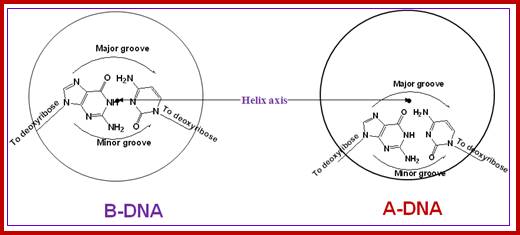
A-DNA and B-DNA; DNA structures are now determined using crystals of synthetic oligonucleotides. This has improved the precision of the description of DNA structure, but there is still some ambiguity about the structure of A’ DNA left and B’ DNA right. Principles of Biochemistry/nucleic acid 1; http://en.wikibooks.org/
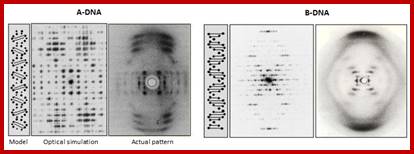
![]() A-DNA
and B-DNA: Comparing their historical X-ray diffraction images;
A-DNA
and B-DNA: Comparing their historical X-ray diffraction images;
A-DNA-The strong features on the 6 to 8th layer lines of both the X-ray picture and the simulated pattern arise from the inclined base pairs seen edge on and forming a zigzagging double slit grating. B-DNA: The strong streaks on the 10th layer lines of both the X-ray picture and the simulated pattern arise from the horizontal base pairs seen edge on.
https://www.quora.com
The most crucial knowledge that Watson gained was, besides the Wilkins and Miss Franklins X-ray diffraction data, the kind of tautomeric forms of nitrogenous bases exist in native state, where adenine and cytosine in amino form (they can also exist in Immino state) and guanine and thymine in ketonic state (which can also exist in enol state); in these states they base base-pairs with each other; this information was provided by Jerry Donohue (Linus Pauling’s Student), a friend of J.D Watson. Chargaff has already provided the information about quantitative analysis of Adenine, Thymidine, Cytosine and Guanine of DNA, where the quantity of Adenine found to be equal and thymidine found equal to Adenine. This information has come as ‘lightening from the blue’ “enlightenment”. The problem they were facing was which base paired with which base. It can be A=A, C=C, G=G, T=T, A=T, A=G, G=C, and G=T. Donohue’s information that A and C exist in amino form, G and T in ketonic form and no other structural forms possible in cytosolic fluid. Chargaff’ (very intelligent) did understand the equivalence of A = T and G = C concentration; but could not come up with the structure, for he was too busy with administration. If you want to spoil a great scientist give him an administrative job. Chargaff was very intelligent and also busy in administration (?) that made the difference between getting Nobel Prize and not getting it but sadly watched the Nobel Prize awarding ceremony to Watson, Crick and Wilkins.
Comparison of DNA Nitrogen bases (Nucleotide Quantities) from different sources:
|
|
A |
G |
T |
C |
A+T/G+C |
|
Human |
30.4 |
19.6 |
19.9 |
30.1 |
1.53 |
|
Rat |
28.6 |
21.4 |
20.4 |
28.4 |
1.33 |
|
Sea Urchin |
28.4 |
19.5 |
19.3 |
32.8 |
1.58 |
|
Carrot |
26.7 |
23.1 |
17.3 |
26.9 |
1.16 |
|
Clover |
29.9 |
21.0 |
15.6 |
28.6 |
1.41 |
|
Neurospora |
23.0 |
27.1 |
26.6 |
23.3 |
0.86 |
|
E.coli |
24.7 |
26.0 |
25.7 |
23.6 |
0.96 |
|
T4-Phage |
32.3 |
17.6 |
16.7 |
33.4 |
1.91 |
Quantitative analysis of nitrogen bases, which showed that the concentration of Adenine was found equal to Thymine and Guanine, was equal to Cytosine, and A+T not equal to G+C. None of the scientists understood it but for Watson it was enlightening and significant, even the person who did work on chemical analysis of Nitrogen bases failed to understand it. It is a travesty of truth for Chargaff.
Among many scientists who were working on DNA, in fact only few were in the race to solve, and the expectations were very high and coveted Nobel Prize was so near and dear, yet it was a distant dream, a tantalizing experience for many. Watson was one among the motley crowd. When the triple stranded DNA model by Linus Pauling was sent for the publication, every one thought the mystery was over and the structure was solved once and for all to usher a new era in the field of biology. Watson never flinched or felt sad that he has lost the race, instead, when he read the contents of the paper, he felt that L. Pauling’s proposed model was somehow wrong, did not fit into scheme of things that he had in his mind, perhaps a smile within himself; it was a desperate moment for him. In spite of Pauling’s overpowering personality, two time Nobel Laureate and knowledge about macromolecules, especially of proteins, and of chemical bonds; it is Dr. Pauling who elucidated alpha helical structure of polypeptide chains, and it is he again discovered Hydrogen bonds and their role in providing stability to the alpha helical structure of polypeptide chains; for which he was awarded a Nobel Prize; in spite of it, it is paradoxical that Pauling could not come to terms and bungled in proposing triple stranded DNA model, perhaps he failed to understand x-ray diffraction studies on DNA or someone deliberately failed to provide him with the correct X-diffraction pictures; in fact he did not get Miss Franklins X-ray diffraction picture of DNA and the data. He was so near, yet far, if he had solved it would have been, a momentous time in the history of biology and with another Nobel Prize, it would have been crowning moment for a great person who richly deserved and it would have been a history of sort in the Nobel Prize academy. Miss Franklin drifted away from the subject and to some other place; unfortunately, she died before the structural elucidation of DNA.
Watson’s insight into the structure was so profound, it required only one simple mental break through about base pairing; which base paired with which base, what was the tautomeric state of each of the bases involved in perfect and unambiguous base pairing, and what chemical bonds, (whether covalent bonds or Hydrogen bonds,) were responsible for correct base pairing, and how many of chemical bonds were involved in each kind and combination of base pairings. To specify each of the bases pairing, as well as provide enough strength to be stable and strong enough to withstand tumultuous events of replication and recombination at molecular level, and how one could explain absolute, precise and exact mode of replication and the information flowed downstream.
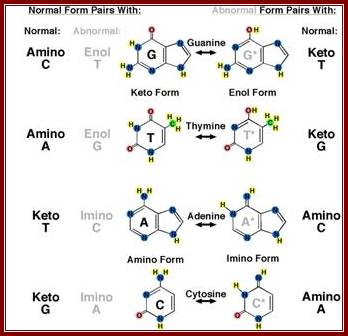
This diagrammatic representation of Nitrogen bases and their possible tautomeric states, where it turned out that Adenine and Cytosine to be in amino form and Thymine and Guanine were found to be in Ketonic state in living forms. A great friend of Dr. J.D Watson provided this tuatomeric form information. This has provided the insights into which base base-pairs with which base and which should provide two hydrogen bonds and which should have three hydrogen bonds. So, Watson got Nobel Prize and his friend got only thanks. http://sandwalk.blogspot.in/ http://eternawiki.org http://eternawiki.org
Arrows show attractive forces that operate on nitrogenous bases of nucleotides
Laurence Moran; http://sandwalk.blogspot.in/
The enlightenment came one day when he was returning to this apartment, it was a flash, and in a flash, he solved, and solved “the jig-saw puzzle” instaneously. Such breakthroughs occur to scientists who work ceaselessly, work day in and day out, sleep and dream of discoveries while they work. Such ‘avant-garde’ discoveries don’t happen to people just by luck. You may publish 1400 or more papers and publish 30-40 books and you may get all sorts of local prizes but not “The Nobel Prize”. There is huge difference between these two.
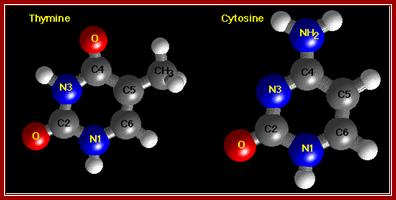
http://chemistry.oregonstate.edu/
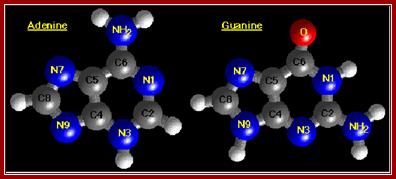
The diagrams show the ball and stick model of Bases. http://chemistry.oregonstate.edu/
When Watson sat down to write the diagrams of base pairs to decipher, he was very clear in his mind that ketonic form of Guanine paired with amino form of Cytosine with possible 3 hydrogen bonds, and amino form of Adenine paired with ketonic form of Thymine with possible 2 hydrogen bonds, and he knew for certain that there are no other combinations possible to satisfy geometrical, stearic, chemical and most importantly thermodynamic parameters. Prof. Chargaff’s quantitative analysis fit with Watson’s concept of base pairing, with Crick to explain the salient features of DNA as the molecule of life or “Mother of Life”. Watson and Crick together proposed the model and published their paper, a par excellent one, as a letter to the editor in “Nature”. But wrong and immediately corrected and sent another paper to nature, in which they corrected their mistake. The paper was short but sweet, it has provided the quintessence of DNA structural features inspite of some lacunae in their proposition. This contribution was an epoch making one in the history of biology and set the seeds for a revolution in molecular biology, a new era of “Molecular Biology” started. The model proposed by Watson and Crick explained basic features, and at later point of time different people have added more information and other structural details. (using this information from centre spread article from “LIFE “, I, the author of this website, gave a lecture in our departmental ‘student lecture competition’ in Central college, Bangalore in 1963; I did not know much of Nucleic acids; since I gave the lecture and won the prize, then our teachers appreciated me and started teaching DNA and RNA under subject Cytology in Bangalore University; till then they did not bother to teach these subjects.” Perhaps I, as a student was ahead of professors at that time point. The same year in my exam questions were asked on DNA and RNA; I was delighted and answered well. J D. Watson was kind enough to send his Book Molecular Biology of the Gene first edition to me; I was so happy you know when you too happy what you do, I did the same.

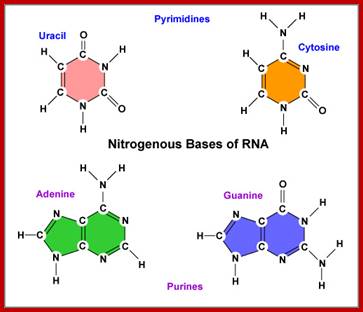
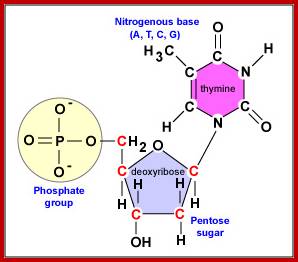
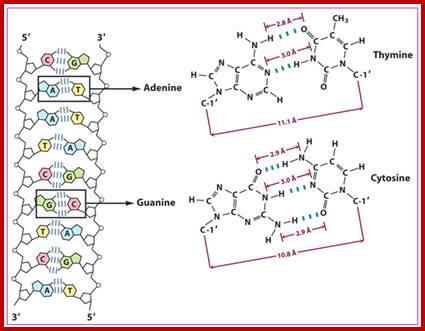
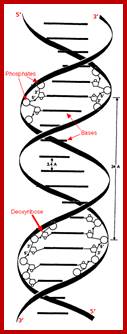
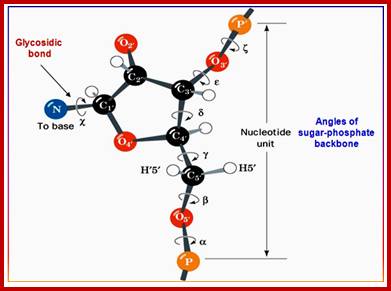
Basic Nucleotide structure with deoxyribose sugar bound to a nitrogenous base at C1and N9 (base). Phosphate bound to Sugar at 5C sugar; In DNA the sugar is Deoxyribose; not ribose. https://www.ncbi.nlm.nih.gov; https://www.researchgate.net
Molecular Genetics; http://faculty.ccbcmd.edu

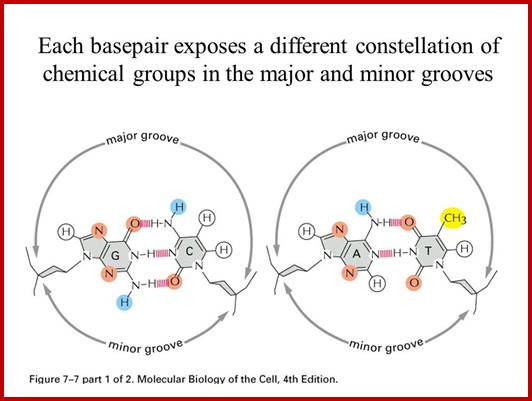
The standard base-pairing between specific Purines and Pyrimidines; Look at the strong G and C pairing; www.studyblue.com and https://quizizz.com
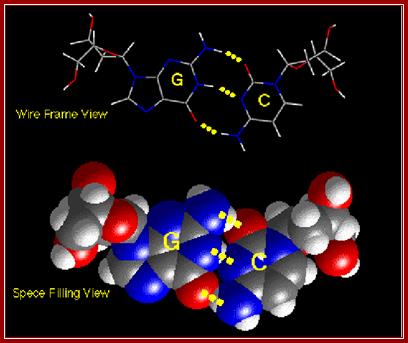
Base pairing between G and C-ball and stick model. http://chemistry.oregonstate.edu/
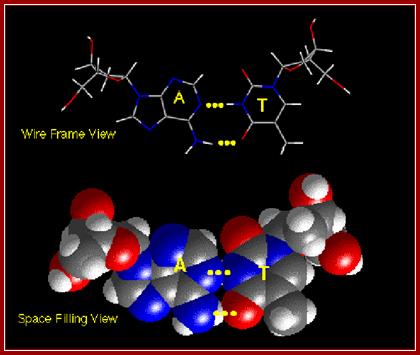
The above diagrams show base pairing between A and T; http://chemistry.oregonstate.edu/

Sugar ring conformational changes; C3’endo and C2’exo pucker the sugar in cis or syn configuration and C2’endo and C3’exo in anti-configuration. http://x3dna.org/highlights
http://x3dna.org/highlights/sugar-pucker

The stereo models represent sugar puckering, such as C’2 endo and C’3 exo in the case of sugar attached to Adenines or Guanines in C1’ positions which provides the Phosphate and the bases in ‘cis’-configuration with respect to their orientation. This happens when the sequence of double stranded polynucleotide chain contains 5’CpGpCpGpCpGpCpG3’ repeats of 5-10 base pair long. The C3 endo and C2 exo produce cis or syn configuration, this configuration happens when the sugar C1 is bound to Thymine or Cytosine; When they base pair in a long polynucleotides strands; their rotation in space with respect central axis is determined; http://x3dna.org/highlights
Understanding of base pairing and the configuration of phosphate group and nitrogen base with respect to ribose sugar is important. Nitrogen bases are hydrophobic in nature and Phosphate group hydrophilic in nature. The structural feature of foldable ribose sugar called ‘PUCKERING” is important. The configuration of nitrogen bases with respect to sugar can be syn or anti. Based on the neighborhood bases the sugar can fold or configure into C2 endo and C3 exo or C2 exo and C3 endo. When C3 endo and C3 exo configuration the nitrogen base is syn where Phosphate and base are fold towards the same side. On the contrary when C3 exo and C2 endo the bases are in anti-configuration. If the base sequences are –GCGCGCGCGCGC- ten or more, the opposite strand also contain the same sequence like –CGCGCGCGCGCG-- in opposite polarity. In such configuration where C3 endo and C2 exo with guanine base, the base folds in cis form with respect to puckered sugar. In a ds stranded DNA when alternate nucleotides with opposite configuration are base paired the P-S-P-S-P-P back bone shows zig-zag contour; and dsDNA takes left hand coiling; on the contrary the random sequences of nucleotides when base pair show smooth curved S-P-S-P-S-P-S-P back bore where each of the nucleotides are in C3 exo and C2 endo configuration with respect to sugar puckering takes right handed coiling. In this flat base, the hydrophobic N2 bases in opposite position twisted to each other, they are not in the same plane, this is true with right handed DNA helices. This configuration generated by such sequences cause the DNA to take left hand coiling or right-hand coiling.
This diagram again represents the C3’ endo and C2’endo puckering with Cis and Anti configurations, from this one can make out which has cis-configuration and which has anti-configuration with respect to N2-baae and Phosphate at each ends of deoxyribose sugar; http://www.siumed.edu/
Antiparallel Strands;’ In a double stranded DNA molecule, the two strands run antiparallel to one another so one is upside down compared to the other. The phosphate backbone is located on the outside, and the bases are in the middle. Adenine forms hydrogen bonds (or base pairs) with thymine, and guanine base pairs with cytosine.
https://www.boundless.com
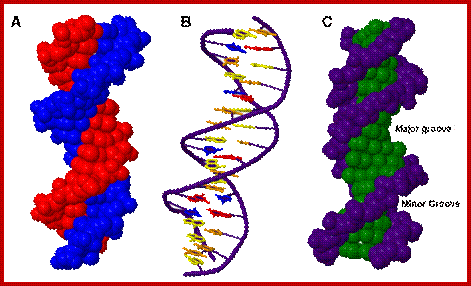
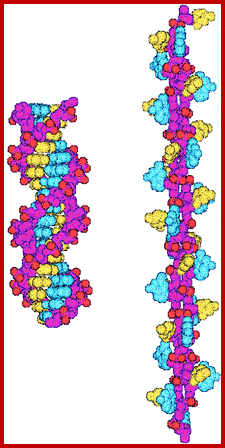
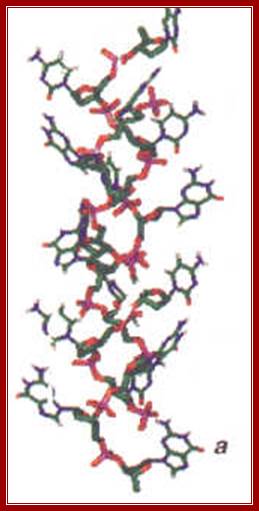
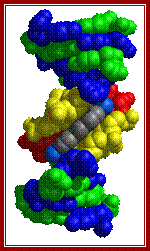
Three representations of DNA's double helical structure. A is a spacefill model of DNA, where every atom is represented as a sphere. The two anti-parallel polynucleotide strands are colored differently to illustrate how they coil around each other. B is a cartoon model of DNA, where the sugar-phosphate backbones are represented as violet strands and the nitrogenous bases are represented as color-coded rings. C is another spacefill model, with the sugar-phosphate atoms colored violet and all nitrogenous base atoms colored green. The major and minor grooves, which wrap around the entire molecule, are apparent as the spaces between the sugar-phosphate backbones. https://www.boundless.com
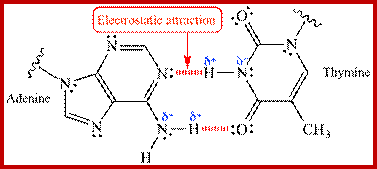
(Electrostatic attraction shown in red “Van der Waals “ interaction between the δ+ and δ- ends of a polar covalent N-H bond allow for hydrogen bonding and base pairing within the DNA double helix; http://www.chem.ucla.edu

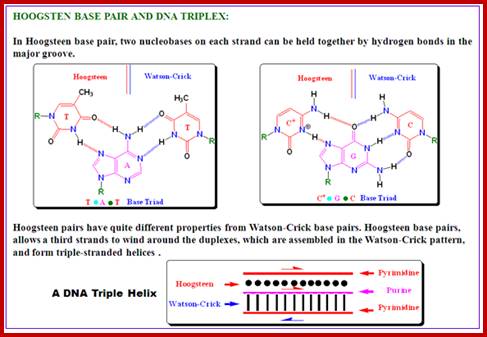
The difference between Watson-Crick and Hoogsteen pairing. https://www.slideshare.net/ (Illustration by Kevin Spear);
http://blog.drwile.com/
http://pubs.rsc.org/
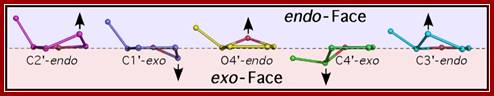
Sugar pucker. Shown are the endo (above) and exo (below) faces of the 5-membered furanose sugar with the nucleotide base extended above the reference plane. Sugars are shown in order of transformation from C2’-endo to C3’-endo. Arrows indicate the atom that is puckered, and the direction of puckering. http://www.intechopen.com/
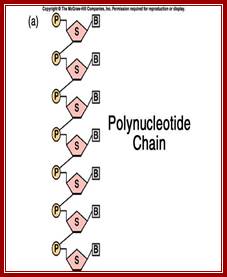
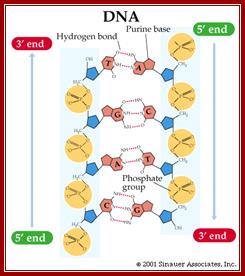
Single stranded polynucleotide chain and double stranded DNA with Watson and Crick base pairing; http://cmgm.stanford.edu/ http://www.angelfire.com/
http://chemistry.oregonstate.edu/
![\includegraphics[width=10cm]{figs/chapter3/secondary.eps}](Deoxy_Ribonucleic_Acid2-Structure_files/image053.jpg)
Stacking is an intermolecular interaction observed in aromatic molecules that tend to arrange them in a pile (see Fig. 3.3b). There are two forces that stabilize base stacking: the hydrophobicity of the aromatic rings of the bases and the London dispersion of the dipoles (induced in the bases). Stacking forces are different for each combination of base-pair. In general, a stack of purines (adenine and guanine) is stronger than a stack of pyrimidines (cytosine and thymine). Apart from Watson-Crick base-pairs, there are also other motifs (such as loops or bulges) that can contribute to the thermodynamic stability of the secondary structure.; Secondary structure of DNA. (a) Hybridization of two antiparallel strands of DNA. The straight lines represent covalent bonds, while the discontinuous ones represent hydrogen bonds. (b) Stacking of four bases. The two strands run in opposite directions. There are two forces that stabilize base stacking: the hydrophobicity of the aromatic rings of the bases and the London dispersion of the dipoles (induced in the bases). Stacking forces are different for each combination of base-pair. In general, a stack of purines (adenine and guanine) is stronger than a stack of pyrimidines (cytosine and thymine). Apart from Watson-Crick base-pairs, there are also other motifs (such as loops or bulges) that can contribute to the thermodynamic stability of the secondary structure. http://www.tesi.mintrada.com
![\includegraphics[width=\textwidth]{figs/chapter3/tertiary2.eps}](Deoxy_Ribonucleic_Acid2-Structure_files/image054.jpg)
Tertiary structure of DNA. (a) A-DNA. The phosphate-deoxyribose backbone is represented in green; adenine in red; cytosine in blue; guanine in orange; and thymine in yellow. (b) B-DNA. (c) Sugar puckering. The upper picture shows the chemical formula of the 2`-deoxyribose. The lower pictures show the two possible sugar pucker conformations. http://www.tesi.mintrada.com
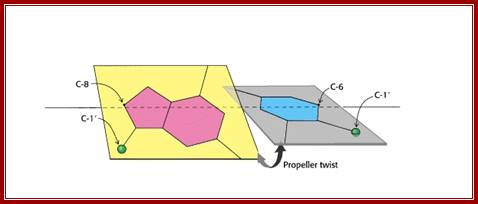
Propeller Twist: The bases in the opposite strands of DNA are often not precisely coplanar. They are twisted with respect to each other, like the blades of a propeller. Propeller twist is higher for A-T than G-C by 5-7 degrees; Hydrogen bonds are not that strong and do allow such variations. Base pairs in ds DNA stack in such a way they exclude water; http://www-personal.umich.edu/ ; http;//cmgm.stanford.edu
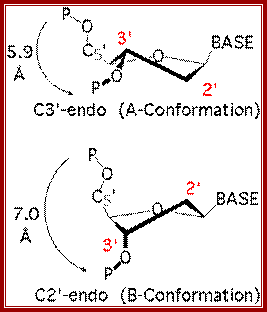
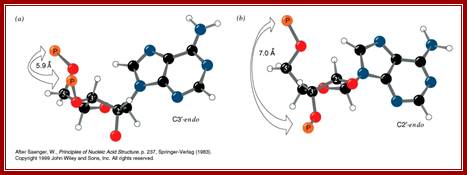
C3′-endo and C2′-endo sugar puckers are highly correlated to the perpendicular distance between the C1′–N1/9 glycosidic bond vector and the following phosphate: > 2.9 Ĺ for C3′-endo and < 2.9 Ĺ for C2′-endo. From Mol Probity; http://haddock.science.uu.nl
www.slideplayer.com

shows base pair stacking in B-DNA. Short range repulsion sets a distance of 3.4 Ĺ between base pairs. http://ww2.chemistry.gatech.edu

DNA base pairing and base-pair stacking induces Propeller twist. https://www.youtube.com

Base Pairs stacking; Biochemistru Medicinalis; https://www.slideshare.net
The ds DNA is more stable for the bases are paired according to their compatibility. The base pairs are flat with hydrophobic forces, where they are not paired as flat but they are slightly twisted to each other in spite of the hydrogen bonds. The strongest stacking is G- C and little weak in A-T. The stacked base pairs do attract to each other by Van der Waals forces from 2 to 4kJ/mole^-1, but the dominant driving force stabilizing base stacking is non-hydrophobic solvent entropy. The stacking force create flexible DNA into curved structure and it coils in right handed direction to be stable; Chi H. Mak; http://pubs.acs.org.
Forces that stabilize DNA are Hydrogen bonds between complementary Bases by electrostatic interactions. Stacking is made-up of two separate forces: hydrophobic effect and London dispersion forces. Hydrophobic base pairs remain free from water. DNA stacking also shows nearest-neighbor interaction.
Stability: Aromatic Stacking - Weak noncovalent force caused by overlapping of p-orbitals; also called pi stacking. In DNA, aromatic stacking between the nucleotides contributes to its stability. The pyrimidine and purine bases, which are parallel to each other in DNA, participate in aromatic stacking due to the overlap of their p-orbitals. - Hydrogen Bonding - Millions of hydrogen bonds in DNA is the main structural feature that explains why DNA is stable. Hydrogen bonding is strong, but can easily be broken for DNA replication.
wwwnptel.ac.in

DNA ds Helix model; http://chemistry.oregonstate.edu/
In 1974, oligo’s containing sequences such as CGCGAATTCGG and CGCGCGCG were synthesized and their crystals were subjected to X-ray crystallographic studies. Overall structural features of DNA crystals, though basically same as propounded by J.D Watson and F.C crick, but few more additional features were added.
The double stranded DNA structure proposed by Watson and Crick is called B-DNA. But of late, based on the sequences and of environs of DNA, it is known that it can assume different structural forms such as Z-DNA and A-DNA. In solution ds DNA helix is a flexible structure, undergoes dynamic conformational changes all along its length. It is constantly subjected to, local but thermally induced fluctuations in the arrangements of its atoms, which cause individual molecules to bend, twist and stretch. Such distortions represent changes in rotational angles of covalent bonds along the polynucleotide backbone. Spontaneous kinks and bends and other transformations require little amount of energy. What is known today about DNA structure is that it can vary with sequences and external forces, so one cannot simply assume DNA has a particular structural form for it is a dynamic structure. Any change at any one position of it will be transmitted all along the length of the helix. DNA shows a variety of structural features such as, ss DNA, ds DNA, ts DNA, 4sDNA, curved or bent DNA, kinked DNA, hairpin or cruciform, relaxed DNA, super coiled form and perhaps another DNA called P-DNA (Peptide DNA) and other forms. It depends upon the sequence context, its association with other components, and concentration of water, pH, temperature and ionic concentrations; certainly, the basic structure is not uniform throughout the length of the DNA; there will be distortions based on the associated proteins and other components.
Stretched and overwound DNA forms a Pauling-like structure with exposed bases; Structure of P-DNA deduced from molecular modeling. Space-filling models of a (dG)18⋅(dC)18 fragment in B-DNA (Left) and P-DNA (Right) conformations. The backbones are colored purple, and the bases are colored blue (guanine) and yellow (cytosine). The anionic oxygens of the phosphate groups are shown in red. These models were created with the JUMNA program (11, 12) by imposing twisting constraints on helically symmetric DNAs with regular repeating base sequences; J. F. Allemande, http://www.pnas.org
B-DNA:
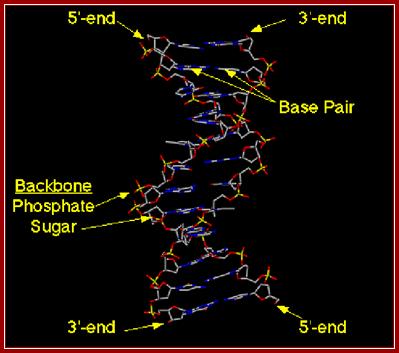
· DNA is made up of 2 polynucleotide strands of opposite polarity (5’-3’).
· The two strands are helically coiled to each other and coiling is in right hand sense.
· The bases, which are flat, rigid and hydrophobic are attached to sugars by glycosidic bonds and oriented in anti configuration with respect to 2’ endo and 3’ exo sugar puckering.
· The bases are projected at right angles to the sugar-phosphate-sugar backbone. The bases are slightly twisted in space with respect to their sugar phosphate axis.
· Two strands are held to each other by specific base pairing by hydrogen bonds, dictated by base’ tuatomeric states. The flat bases are paired not as straight flat structure or what is called co-planar, but in specific tilted angles to each other. This tilt is torsional. This is the basis for DNA twist.
· The base pairs which are hydrophobic, having a geometry of their own, are not co-planar to each other but stacked one above the other; but because of hydrophobic interactive forces make stacking one above the other energetically not compatible, thus the hydrophobic forces drive stacking partially overlapping and thus generate a kind of propeller twist, which in fact drives the helix in right handed direction.
· Base pairing between A=T and G=C provides specificity and stacking interactions (hydrophobic) provide stability.
· In the double helix, the sugar phosphate backbone is not extended as straight structures, but sugars inwardly puckered to outwardly projected phosphate groups. In sugar –phosphate backbone, 5’ P – 5’ C – 4’ C – 3 ‘ C – 3 ‘ P, the torsional angle between 4 ‘C and 3’ C is restricted and constrained, but there is certain amount of freedom of angle of rotation between 5’P to 5’C to 4’, similarly one finds such freedom of rotation at 3’ end that is between 3’ C - p – 5’C. In spite of such constraints the double helix is reasonably strain free.
· The coiling excludes water molecules from the inner space of the coil, because the base pairs are hydrophobic in nature.
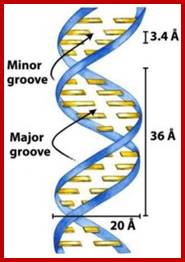
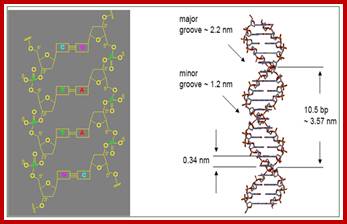
· The diameter of the helix is approximately 20 A°, and the distance between to nucleotides is about 3.4 A°. The angle of twist is ~3.6 degree per the raise of each pair of nucleotides. Thus, for every 10.5 to 10.6 base pairs the DNA takes one complete right-handed turn. The pitch raises per turn of 3.4A° (the pitch rises per turn). The aromatic bases have Vander Waals thickness of 3.4 Ĺ
· Because of the two antipolar strands that helically twisted to each other, with bases projecting inwards into the helix, DNA structure shows two defined spaces called grooves, one of them is called major groove, which is broader (22 Ĺ) and deeper, while another groove that generates because of two strands strung together is called minor groove of 12 Ĺ diameter.
· The geometry of DNA structure is entropically favored and thermodynamically favored but enthalpically apposed. Watson-Crick base pairing is the most favored electronic complementarity. Any other form of base pairing is called non-Watson-Crick base pairing (another alternate form of base pairing is called Hoogsteen Base pairing) and such base pairing is considered as possible under certain situations where non-Watson-crick base pairing is often forced.
· The strength and the stability of the helix mainly stems, not from hydrogen bonding, but from hydrophobic interactions between base pair stacking. Hydrogen bonding contributes to specificity of base pairing.
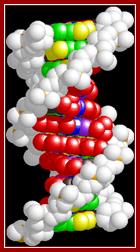
ds DNA wire model; http://chemistry.oregonstate.edu; and http://www.bic.arizona.edu.
· Non-polar solvents like ethanol should actually strengthen hydrogen bonding than water, yet the DNA structure is destabilized by ethanol, because ethanol disrupts hydrophobic interactions, so it is the hydrophobic interactions between the base pair stacks that provide the strength.
· Water provides the favored environment for the structure of the DNA and also provides the forces that contribute to its stability, for example cationic salts by binding to negative Phosphate ions do add to the stability of the DNA structure.
· Stability of DNA structure is enthalpically driven and entropically apposed, while protein structures is entropically driven and enthalpically apposed

biochemia.wp.uph.edu.pl/ B-DNA helix; http://luthervandrosss.blogspot.com

http://pubs.rsc.org
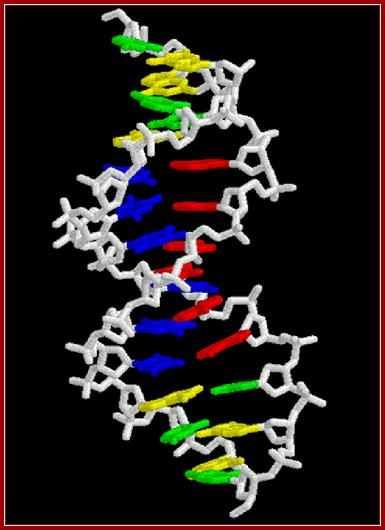
www.biochemia.wp.ap.siedlce.pl;B DNA another model-it is joy look at these models

www.biology.arizona.edu
Z-DNA:
Isoform concept is factual one, so when a right-handed structure exists there is always the possibility of left-handed structure. The concept of left-handed DNA was not a figment of imagination. The concept of left handed structure took its seeds in Indian Institute of Science, Bangalore, in Prof. G. N. Ramachandran’s lab; he was an erudite par excellent scholar, the ‘great’ among Indian Molecular Biologists.
Its existence was soon realized. Various thermodynamic studies using various combinations of sequences revealed that left-handed structure as a possibility. Now it is unambiguous that a left-handed DNA structure is a reality when a sequence of CGCG is repeated more than two to three times, with no other constraints acting upon, the segment takes a left-handed helicity. This is possible for the G’s in such sequences are in cis configuration (puckering) with respect to their sugar conformation, which because of the constrained sequence, forced to assume puckering of 2’ exo and 3’ endo. But the pyrimidine nucleotide exists in anti configuration. Such alternate cis and anti configured of sugars in nucleotides with their bases, forces the sugar phosphate backbone to assume a zig-zag conformation. The inwardly projected twisted bases in anti and syn conformation drives the helix into left-handed turn. The helix does not have smooth contour of sugar-phosphate-sugar backbone as found in B-DNA, but it has a zig-zag course. In spite of drastic conformations, base pairings and the overall structure is more or less same with certain exceptions, such as thickness of the helix, size, angle of rotation, rise per nucleotide and rise per helical turn and depth of major and minor grooves.
Crystal Structure of Hexameric Z-DNA:
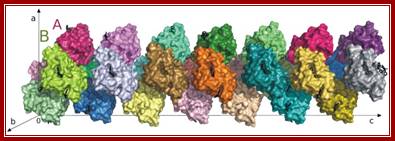
The crystal structure of a Z-DNA hexamer has been solved from the anomalous signal of the P atoms at copper wavelength. The multiplicity of the diffraction data was the most crucial single factor for the solution of the phase problem. The structure was refined to an R factor of 0.089 at 0.95 A resolution. In another study, we have demonstrated the power of the (Ta6Br12)2+ complex for phasing protein structures and determined the precise geometry and molecular interactions of this cation in a protein crystal context. The crystals of Hyp-1 in complex with ANS turned out to have commensurately modulated superstructure with 28 protein molecules in the asymmetric unit. The structure of this crystal (which also showed tetrahedral twinning) was solved (in collaboration with Prof. Randy Read, Univ. of Cambridge) by Molecular Replacement despite extremely complex, seven-fold, translational Non-Crystallographic Symmetry (tNCS). Modulated crystal structures are extremely rare in macromolecular crystallography. We are also developing Internet tools for teaching crystallography, available at this link (including PXQuiz); http://www.man.poznan.pl/CBB/research.htmlhttp://www.man.poznan.pl/CBB/research.html
· It consists of two polynucleotide strands of opposite polarity.
· They are helically coiled to each other, but the direction of the helicity is left-handed.
· The left- handedness stems from the alternate bases in cis and anti conformation. The base pairings are always between a cis configured base in one strand with anti configured base in the opposite strand.
· The base pairing however is again between G & C, driven by tuatomeric state of bases as complying with Watson-Crick base pairing.
· The propeller twist that is generated because of this alternate anti and cis sugar configuration and base pair stacking with zigzag course of sugar-phosphate backbones the helix takes left handed helicity.
· Such Z kind of conformation is more easily driven into, if the cytosine residues are methylated at C’ residues of CGCGCGCG sequences; furthermore, if such sequences are bound by sequence specific DNA binding proteins; and such DNA is further stabilized. Such structures are found clustered within the long stretches of B-DNA.
· Many such CGCG clusters or islands and their locations have been identified in many genomic DNA. And surprisingly they are found in the promoters or in the vicinity of promoters.
· This kind of configuration is restricted to only short stretches where G=C sequence prevails.
· Such regions in Z-DNA help the DNA in unwinding either during replication or transcription.
· Now it is possible to identify Z-DNA and differentiate it from B-DNA segments by using antibodies against Z-DNA and B-DNA sequences.
· The diameter of the helix is 18 Ĺ and the number of base pairs
per turn is 11.
· Over relaxed- longer and thinner, left handed helix.
· Purine C3 endo and C2 exo; Pyrimidines C2 endo, C3 exo.
· Over relaxed and thinner, zig-zag,
· In this stretch of GCGCGCGC, G of one strand shows cis configuration, and the C shows anti configuration, but in the opposite strand where G with cis, base pairs with anti C.
G (cis) = C (anti),
C (anti) = G (cis); this kind of configuration of nucleotides in opposite strand leads to left handed helix with zig-zag sugar-base-phosphate backbone.
Z-DNA –a model; Left handed helix? https://www.dreamstime.com
A-DNA:
A-form;(-http://www.tulane.edu/)
- Most RNA and RNA-DNA duplex exist in this form,
- Shorter, wider helix than B.
- Deep, narrow major groove not easily accessible to proteins,
- Wide, shallow minor groove accessible to proteins, but lower information content than major groove.
- Favored conformation at low water concentrations,
- Base pairs tilted to helix axis and displaced from axis,
- Sugar pucker C3'-endo (in RNA 2'-OH inhibits C2'-endo conformation),
· When the B-DNA is subjected to a dry condition, say 75% of water, the B-DNA assumes a structural form that is squatter, thicker and wider having 11.2 bp per turn, which is right-handed.
· RNA-DNA hybrids do show A-conformation.
· RNA-RNA hybrid molecules also show A conformation.
· Hairpin or stem loop structures show A- conformations.
· Sugar puckering in A DNA is 3’endo.
‘A’ DNA- Model-?
Properties of three forms of DNA: http://www.angelfire.com/

Water –hydration in the minor Groove-(d(CGCGAATTCGCG)] 2 (AATT); http://www1.lsbu.ac.uk
Hydration structures in the minor grooves of the unmodified (a), DD1a (b), DD1b (c) and DD2 (d) DNA: DNA duplexes. The unmodified duplex is shown in blue lines while the present duplexes are shown in red lines. The aminohexyl, carbamoyl and methoxyl groups are colored green. In the unmodified duplex, the cyan spheres are water molecules and the gray spheres are solvent molecules partially occupied by sodium ions and water molecules. In DD1a and DD1b, the water molecules are in cyan, and the potassium ions are in gray:
https://openi.nlm.nih.gov
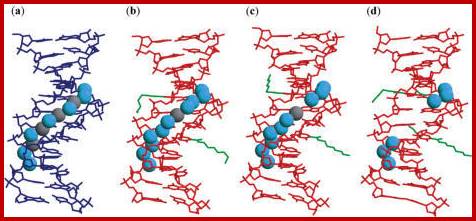
https://openi.nlm.nih.gov

Cross sectional view of DNA: http://mu-peter.blogspot.com/
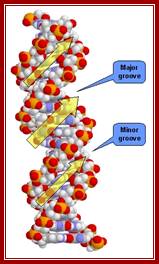
The major and minor grooves form between the phosphate backbones of the DNA molecule, so that the obtuse angle forms the major groove, and the acute angle forms the minor groove.
http://tonga.usp.edu
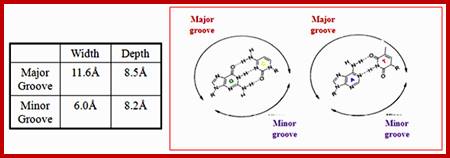
Groove dimensions and the WC base pairs; Helical structure repeats every 34A; Major and minor groove are the two surfaces that wind about the outside of the helix. They are formed by the edges of the stacked bases. These grooves are distinct because they have different H-bonding patterns and different size. Major and major groove of DNA contain sequence-dependent patterns of H-bond donors and acceptors. Sequence-dependent duplex structure (A, B, Z, bent DNA); A-DNA has a shallow minor groove and a deep major groove: http://nptel.ac.in;
http://nptel.ac.in/courses
The grooves are formed by the edges of stacked bases and have different sizes because deoxyribose residues are attached asymmetrically (not 180 degrees). The minor groove is the one in which C-1’ –helix axis’c1’ angle is less than 180 degrees (recall that the helix axis passes trough the middle of each base pair in B-DNA) and the major groove is on the opposite side of each base pair.
A/T sequences have a higher negative charge while G/C sequences have higher positive potential. H-bonding to base edges in the minor groove can be used to distinguish between A:T and G:C base pairs. A:T is distinguished from a T:A base pair through indirect read out.
DNA binding drugs are generally flat and small, which makes them fit well into the minor groove. Groove width varies with sequence; A/T rich tracks tend to make the groove width narrower.
The convex shape of the minor groove floor complements the typical shapes of minor groove binding drugs. A/T sequences result in a smooth convex curve whereas G/C sequences have “little” bumps due to the 2-amino groups of guanines.
http://nptel.ac.in/courses
The conformation of a nucleotide unit defined by the seven torsion angles. Double-stranded DNA has limited structural complexity compared to proteins (only 4 nucleotides vs. 20 amino acids). Few secondary structures, no tertiary or quaternary structures.
RNA has some well-defined tertiary structure. Conformation of a nucleotide is specified by 6 torsion angles of the sugar phosphate backbone and the torsion angle that describes the orientation of the base about the glycosidic bond (7 totals). Despite 7 degrees of freedom per nucleotide, they have restricted conformational freedom. http://nptel.ac.in/courses.

A regulatory protein Antp bound to major groove. www.caralgue.com
DAPI a dye (grey) bound to the minor groove of DNA.www.man.poznan.pl
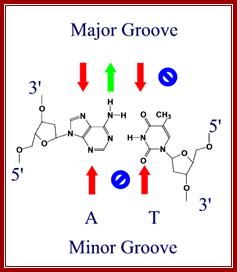
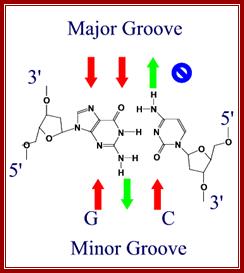
Red arrows indicate attractive forces for Hydrogen bonds and green arrows indicate Hydrogen bond donors; N and O with double bonds provide attractive forces for hydrogen bonding and NH+H groups act as h donors; http://bioweb.uwlax.edu/
http://nptel.ac.in/courses
Cross sectional view of DNA helix: with major and minor grooves providing attractive sites and forces for the binding of proteins;
Bridged nucleic acids (BNAs): DNA conformation is determined by economics in the hydration of phosphate groups.
Bridged nucleic acids (BNAs) are molecules that
contain a five or six-membered bridged structure with a “fixed” C3’-endo
sugar puckering (Saenger 1984). BNA monomers can contain a five-to six-membered or
even a seven-membered bridged structure with a “fixed” C3’-endo
sugar puckering. The bridge
is synthetically incorporated at the 2’, 4’-position of the ribose to afford a
2’, 4’-BNA monomers. The monomers can be incorporated into oligonucleotide
polymeric structures using standard phosphoamidite chemistry. BNAs are
structurally rigid oligo-nucleotides with increased binding affinities and
stability. In crystal structure
analyses of oligonucleotides, the free oxygen atoms of adjacent phosphate
groups along the polynucleotide chain in B-DNA are found at least 6.6 Ĺ apart
and individually hydrated4 whereas they are as close as 5.3 Ĺ
in A-DNA and 4.4 Ĺ in Z-DNA, and bridged by water molecules. We suggest that
this more economical hydration in A- and Z-DNA compared with B-DNA is the
underlying cause of B ![]() A
and B
A
and B ![]() Z
transitions. Use of BDNA= ideal for
detection of short RNA and DNA targets, They high thermal stability of duplex
molecules. They have high stability if they are triplexes and resistance to
exo and endo nucleases, they can be used for strand invasion. https://www.nature.com.
Z
transitions. Use of BDNA= ideal for
detection of short RNA and DNA targets, They high thermal stability of duplex
molecules. They have high stability if they are triplexes and resistance to
exo and endo nucleases, they can be used for strand invasion. https://www.nature.com.

The incorporation of BNAs into oligonucleotides allows the production of modified synthetic oligonucleotides with:
I. Equal or higher binding affinity against an DNA or RNA complement with excellent single-mismatch discriminating power,
II. Better RNA selective binding,
III. Stronger and more sequence selective triplex-forming characters,
IV. Pronounced higher nuclease resistance, even higher than Sp-phosphorthioate analogues,
V. Good aqueous solubility of the resulting oligonucleotides when compared to regular DNA or RNA oligonucleotides.
BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a “fixed” C3’-endo sugar puckering. The bridge is synthetically incorporated at the 2’, 4’-position of the ribose to afford a 2’, 4’-B DNA monomers. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoamidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability. They distinctly different from Peptide Nucleic acids (PNA) and Locked Nucleic acids (LNA).
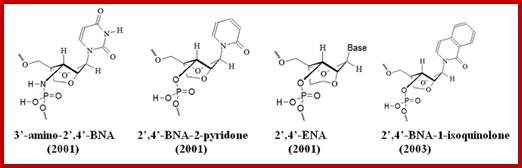
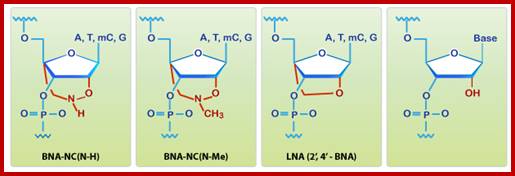

http://www.biosyn.com
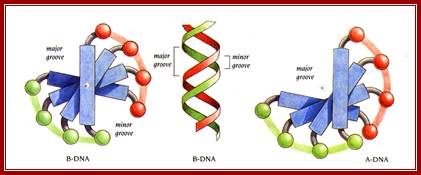
Helix showing the Major and minor groove surfaces
And Base pair stacking and the Propeller twist; http://bioweb.uwlax.edu/
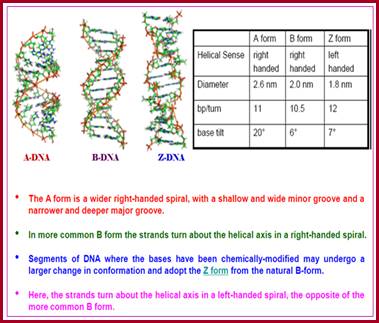
Comparative features of A,B and Z DNA modules;
|
|
A |
B |
Z |
|
Helix nature |
Compact-stout, broader |
Relaxed, longer |
Over-relaxed, longer and thinner |
|
Helical sense |
Right |
Right |
Left |
|
Base pairs per Turn |
11 |
10.5 |
12 |
|
Diameter of the Helix (in A^o) |
26A^o |
20A^o |
18A^o |
|
Rotation per BP (in ^o degrees) |
33.6 |
35.9 |
-30 |
|
Helix pitch (A^o) per turn |
24.6 |
33.2 |
45.6 |
|
Rise per base pair in A^o |
2.3 |
3.32 |
3.8 |
|
|
Major Groove |
Narrow, deep |
Wide, 22A^o, intermediate depth |
Flat, shallow |
|
Minor Groove |
Very broad/shallow |
Narrow, intermediate, 12A^o |
Very Narrow, deep |
|
Sugar Puckering in general: both Pyrimidines and purines
|
C2 endo, C3 exo
|
C2 endo, C3 exo
|
Purines C3endo C2 exo, Pyrimidines-C2 endo, C3 exo |
|
Glycosidic bond orientation with respect to sugar |
Anti |
Anti |
Anti at C and syn at G |
|
|
P to P distance (A^o) |
5.9 |
7 |
7 ? |
|
|
P-S-P backbone |
Smooth |
Smooth |
Zig-Zag |
|
|
Propeller twist (in ^o) |
+18 |
+16 |
~0 |
|
|
Moisture state |
75 % |
92 % |
- |
|
|
Vertical rise (in A^o) |
2.7 |
3.4 |
3.7 |
|
|
Poly-A chain |
A form |
- |
- |
|
|
Poly-T chains |
- |
B form |
- |
|
|
Poly-CG chains |
- |
- |
Z form |
|
|
DNA hair pins |
A form |
- |
- |
|
DNA-RNA hybrids |
A form |
- |
- |
|
RNA-RNA helix |
A form |
- |
- |
Note-Other form of helix is a possibility-such as F, Q, U, V and Y- all synthetic forms, not observed in native state.
|
Structural features of the three major forms of DNA |
|||
|
Geometry attribute |
A-DNA |
B-DNA |
Z-DNA |
|
Helix sense |
right-handed |
right-handed |
left-handed |
|
Repeating unit |
1 bp |
1 bp |
2 bp |
|
Rotation/bp |
32.7° |
35.9° |
60°/2 |
|
bp/turn |
11 |
10.5 |
12 |
|
Inclination of bp to axis |
+19° |
−1.2° |
−9° |
|
Rise/bp along axis |
2.3 Ĺ (0.23 nm) |
3.32 Ĺ (0.332 nm) |
3.8 Ĺ (0.38 nm) |
|
Pitch/turn of helix |
28.2 Ĺ (2.82 nm) |
33.2 Ĺ (3.32 nm) |
45.6 Ĺ (4.56 nm) |
|
Mean propeller twist |
+18° |
+16° |
0° |
|
Glycosyl angle |
anti |
anti |
C:
anti, |
|
C3'-endo |
C2'-endo |
C:
C2'-endo, |
|
|
Diameter |
23 Ĺ (2.3 nm) |
20 Ĺ (2.0 nm) |
18 Ĺ (1.8 nm |
From WIKIPIDIA
|
A form |
B form |
Z form |
|
||
|
Helical sense |
|
||||
|
Right handed |
Right handed |
Left handed |
|
||
|
|
|||||
|
Diameter |
|
||||
|
26 A |
20 A |
18 A |
|
||
|
|
|||||
|
Base pairs per helical turn |
|
||||
|
11 |
10.5 |
12 |
|
||
|
|
|||||
|
Helix rise per base pair |
|
||||
|
2.6 A |
3.4 A |
3.7A |
|
||
|
|
|||||
|
Base tilt normal to the helix axis |
|
||||
|
200 |
60 |
70 |
|
||
|
|
|||||
|
Sugar pucker conformation |
|
||||
|
C-3’ endo |
C-2’ endo |
C-2’ endo for pyrimidines and C-3’endo for purines |
|
||
|
|
|||||
|
Glycosyl bond conformation |
|
||||
|
Anti |
Anti |
Anti for pyrimidine and syn for purines |
|
||
|
Difference between the electrostatic potentials of the three major forms of DNA |
|||
|
A-DNA |
B-DNA |
Z-DNA |
|
|
Negative Potential |
Massively present throughout the global structure but concentrated most highly in the major groove |
Massively present throughout the global structure and evenly concentrated in the major and minor grooves |
Massively present throughout the global structure and present most highly in the minor groove |
|
Positive Potential |
Very sparsely present scattered in the minor groove and hardly at all in the major groove |
Very sparsely present scattered throughout the global structure, mostly in the major groove |
Scattered sparsely in greater amounts than in A-DNA or B-DNA, but outside of the minor groove |
|
Neutral Potential |
Moderately present throughout the structure and mostly in the minor groove, but not in the major groove |
Moderately scattered throughout the structure, but in a smaller amount than in A-DNA |
Moderately scattered throughout the entire global structure apart from the minor groove |
Note:
Helix sense: Refers to helical rotation of a double helix.
Axial rise: The distance between two adjacent base pair stacks. B-DNA shows 3.4 A.
Helix pitch: Length of one complete turn of a DNA is 10.5 A^o in B form.
Base pair tilt: Refers to the angle of flat bases with respect to ds helical axis.
Propeller twist: The base pairs are not perfect coplanar, but, in paired structures each flat base has slight twist or roll angle to each other, which looks like a propeller.
Rotation per residue: Refers to the angle between two adjacent base pairs, in B-form it is 34.3 ^o where the base pairs per turn is 10.5.
Charge Potential: Most A, B anf Z DNAs have negative electro potential.