Types of Damages and Effects:
Living systems have perfected genetics of inheritance. This is achieved
by variety of natural process that leads to natural selection and speciation,
thus the genetic system of an organisms is inherited and perpetuated for
many thousands or even million years, occasionally make mistakes
because variety of causes; such mistakes can happen at gene level
or at chromosomal level, even at cytoplasmic level, means organelle DNA
can suffer damages. Often in course of time these changes if remain
and survived they lead to variation in plants and animals.
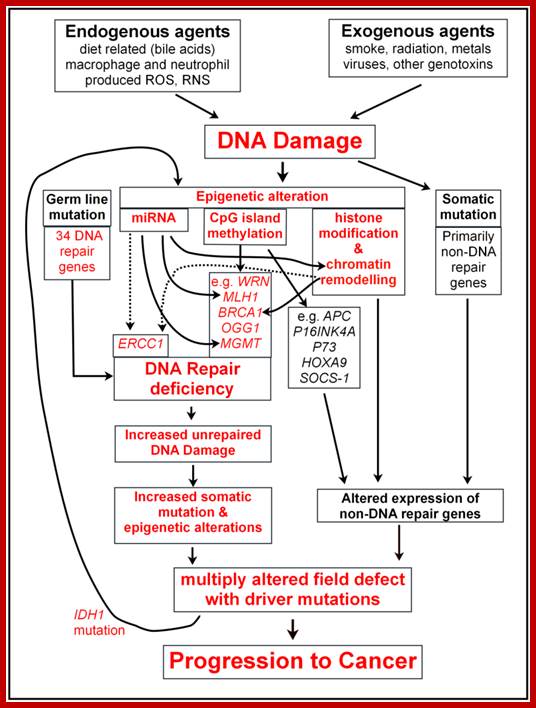
Carol Bernstein, A. R. Prasad et al; www.intechopen.com

Carol Bernstei, A. R.Prasad etal; www.intechopen.com
Chromosomal changes:
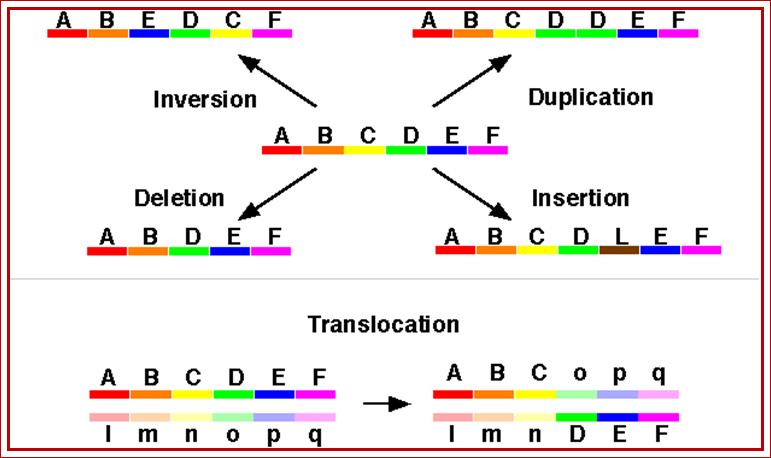
Chromosomal aberrations, as the above diagrams depict, can cause very serious
effect but also lead variations and speciation. http://www.tutorhelpdesk.com/
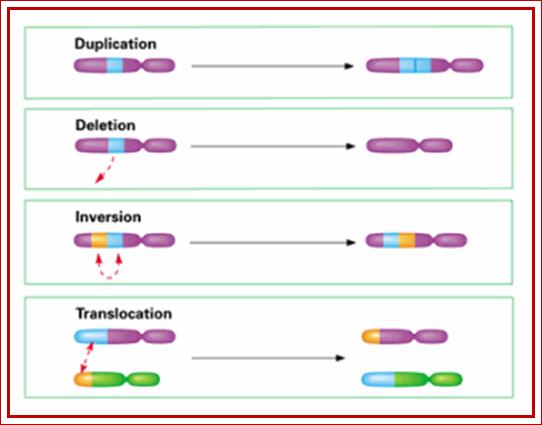
http://www.studyblue.com/
Chromosomal changes can be at numerical level or structural level:
Numerical level can be detected as aneuploidy and polyploidy.
Structural abnormalities can be deletions, duplications,
inversions and translocations. These changes have enormous
effects on organisms, some survive and others perish in the
struggle for existence, those survive inherit their characters differently.
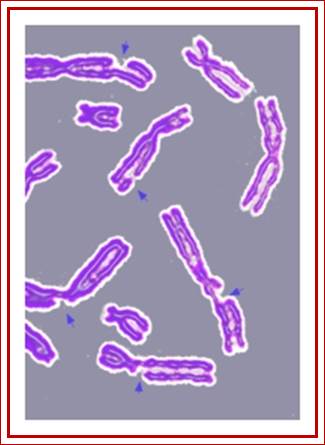
Here is simple chromosomal diagram showing the broken nature of chromosomes. http://en.wikipedia.org/
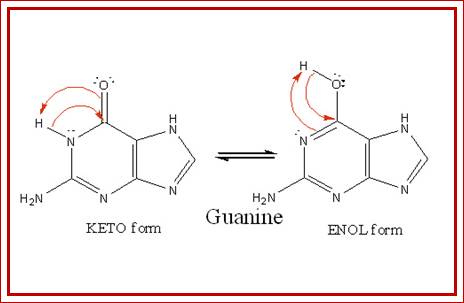
Tautomerization:
Tautomerization involves nitrogenous bases; it is one of the common causes
of genetic errors. Bases under certain conditions can be in a state of enolic
form or immino form instead of their natural ketonic and amino forms.
Such tuatomeric forms can base pair with different bases, which leads to
change in the base pair sequences and generate mutations such as frame
shift in reading frame, silent mutation and nonsense and missense mutations.
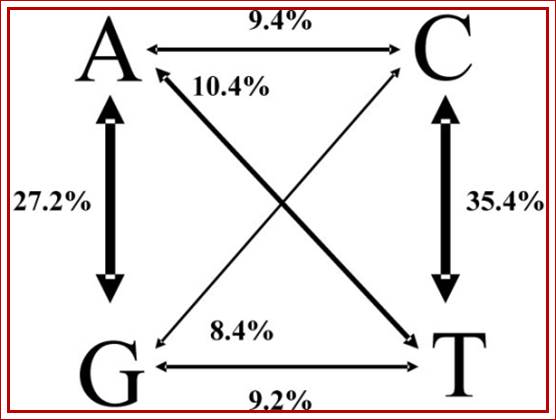
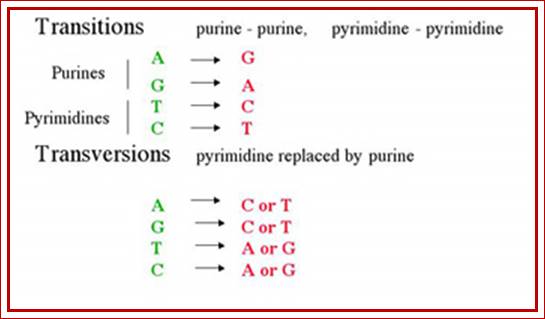
Distribution of Transitions and Transversions among SNPs; A to G i.e one purine or pyrimidines to another purine or pyrimidines; change of
one purine to pyrimidine or pyrimidines to purine (rare), called transversions and transitions respectively. http://openi.nlm.nih.gov/
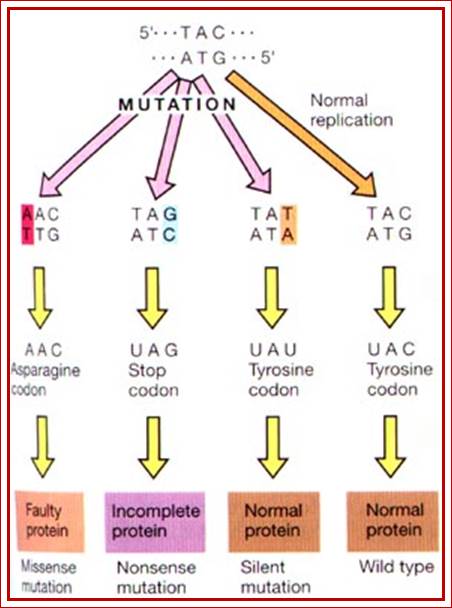
Look at the original triplet sequences, look at mutations- Point mutations can create silent mutations, missense mutations, nonsense mutations as shown above. http://utminers.utep.edu/

Point Mutations; http://www.studyblue.com/
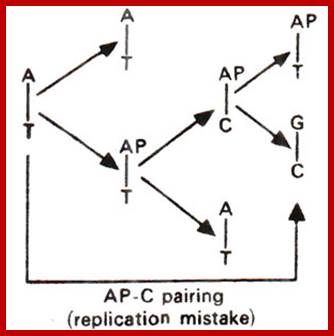


https://en.wikibooks.org

https://en.wikibooks.org
Thymine

Adenine
Effect of tautomerization, where a normal base with either ketonic
or amino form can change into enolic or imminoform; these changes lead to different
base pairing; structural biochemistry; http://en.wikibooks.org/
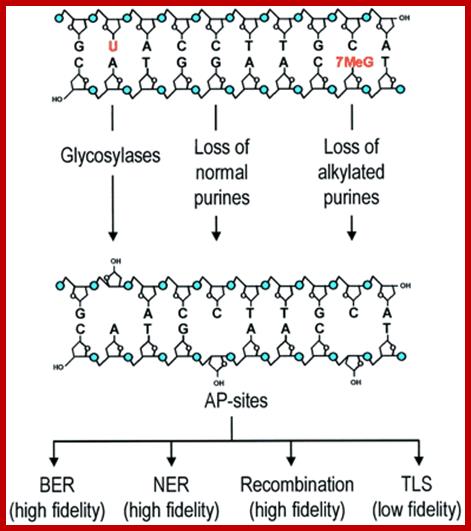
AP sites are mainly generated by DNA glycosylases and depurination. Certain major alkylation products (e.g. 7-methylguanine and 3-methyladenine) increase rates of hydrolytic depurination by several orders of magnitude. Essentially error-free repair of AP sites is carried out by BER, NER and recombination repair, whereas error-prone DNA polymerases may synthesize DNA over the baseless site. This type of complexity in handling of damage may be more common than thought previously. ;https://www.researchgate.net/
· Transitions are due to substitution of purine with another purine and pyrimidine with another pyrimidine and substitution of one purine with a pyrimidine and vice versa invariably due to base substitutions and Tautomerisation. Cellular environment can easily cause such transitions. Even certain type of base modifications can cause transition type changes.
Very rarely Purines are replaced with Pyrimidines or Pyrimidines are replaced with Purines and such changes are called as Transversions.
Such Transversions type incorporations are possible due to mistakes during replication or bases are deleted and new base is added.
Enol form of guanines can easily methylated, which further cause more malformation in terms of information flow. The O-6methyl guanine can base pair with Thymidine,
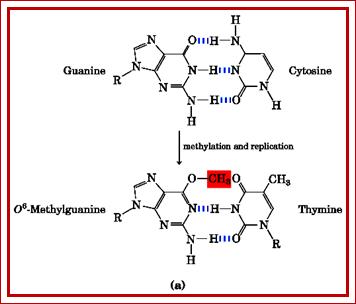
When guanine gets methylated it can base pair with thymine; http://fbio.uh.cu/
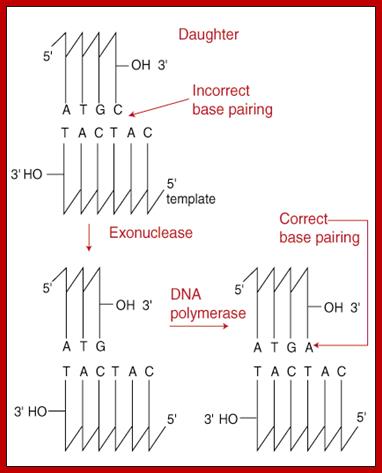
Such changes can be due changes during replication are corrected; 3' to 5' Exonuclease Action If synthesis occurred in the opposite direction, the terminal end of a growing chain would contain a triphosphate group instead of an -OH group. This triphosphate would become the target of the proof-reading exonuclease and its removal would halt DNA replication. http://www.sparknotes.com/biology
In fact replication mechanism and the enzymes involved have inbuilt safety system in the form of proof reading, yet occasionally, perhaps one in one million nucleotides incorporation, there is a possibility, plausibility or probability for wrong nucleotide incorporation, which is enough to create an error. If the error happens in repetitive DNA or transposons does not result in any meaningful changes. If that error in the coding sequences is not repaired or corrected immediately and if the incorporated nucleotide in the codon codes for a wrong amino acid, which if it is important for protein function, such as binding site or catalytic site or both in enzymes or in structural proteins, then the error is manifested in biochemical dysfunction.
Whether it is a prokaryote or a eukaryote, they have inbuilt enzymes with 3’->5’ exonuclease activities, such as DNA pol-I in PK and DNA pol-delta, even DNA pol-gamma have 3’->5’ exonuclease activity, so they perform proof reading, which is called co-replication repair or proof reading mechanism; may have post replication repair mechanism.
- In spite of the presence of such co-replication proof reading activity, incorporation of wrong nucleotides may escape the vigilance of molecular proofreaders. Such error prone incorporations may appear in either of the strands as mis-match nucleotides.
- In prokaryotic systems, there are at least one hundred genes that are involved in repairing DNA damage. Many of them have been identified and their characters and their roles have been delineated.
Prokaryotes are more amenable for creating and detecting single gene mutations than eukaryotes, it is easy to identify and isolate such mutants and genes involved in repair system.
- Eukaryotes also have more than one hundred and thirty or more genes responsible to take care of DNA damage repair system. Yeast, being a unicellular system, has yielded more information about the genes involved in DNA repair mechanism than any other system.
In higher organisms, the DNA damage repair systems are more or less on the same pattern as one finds in E.coli.
- Though it is difficult to obtain mutants in higher eukaryotes, single cell line cultures have provided reasonably some logistic information about DNA repair system, yet but not adequate.
Replication errors:
Though both PK and EK systems have foolproof mechanism of replication, occasional incorporation of non-complementary nucleotides is a possibility, but the enzymes or enzyme complexes that operate have specific subunits with 3’->5’ exonuclease activity. In addition, they have gap filling functions by polymerase functions. In spite of these functions, incorporation of wrong nucleotides does happen, but at a very low frequency, and they escape from the molecular proof reading enzymes called molecular vigilante.
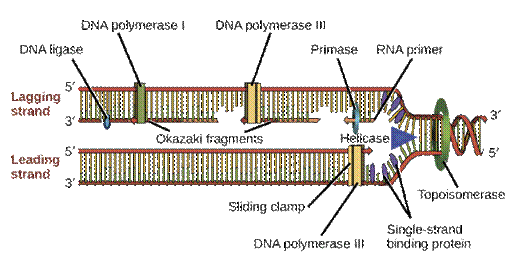
Replication error-AC mismatch; DNA proofreading and repair
Mechanisms to correct errors during DNA replication and to repair DNA damage over the cell's lifetime. https://www.khanacademy.org
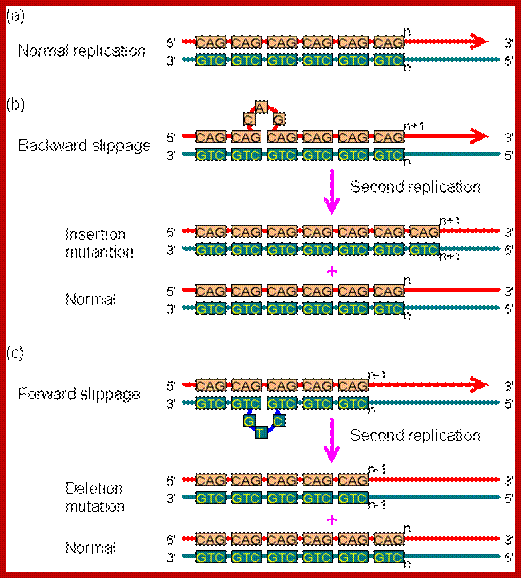
Slippage error during replication; The mutation caused by replication slippage. In this figure, mispairing involves only one repeat. In fact, the slippage could cause several repeats to become unpaired. (a) Normal replication.(b) Backward slippage, resulting in the insertion mutation. (c) Forward slippage, resulting in the deletion mutation. http://www.web-books.com/MoBio
To combat these mismatch errors, cells are endowed with enzymes and the required factors that remove or correct the errors post replication stages and restore the normalcy. Replication can also produce what is called slippage error leading to duplication or deletion of some segments DNA. This can affect the information in terms of reading frame.
Recombination errors:
During recombination, whether it is homologous or site-specific process, occasionally, recombination leads to errors such as loss of certain regions of the DNA or gain in some segments of DNA, which leads to the change in the sequence of the reading frame. This can also produce substitutions, single base deletions and single base additions; all of them generate wrong reading frame, which may work or may not. Such recombination errors are also made during somatic recombination events. Recombination at repetitive DNA often creates mutations.
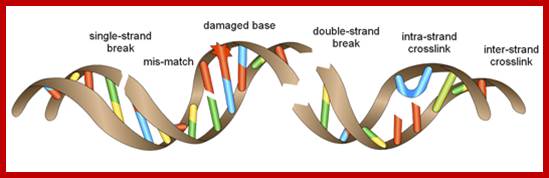
DNA break and repair; http://www.bioch.ox.ac.uk
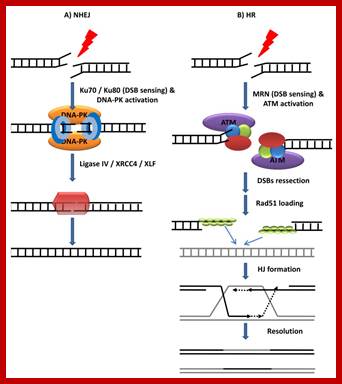
DSBs repair mechanisms. A) NHEJ. DSBs are sensed by the ring-shape heterodimer Ku70/Ku80 which then stabilizes the two DNA ends and recruits DNA-PK. Next, DNA-PK phosphorylates and activates the NHEJ effector complex (ligase IV/XRC44/XLF) that finally religates the broken DNA. B) HR. The ATM kinase is recruited to DSB via an interaction with the MRN (Mre11-Rad50-Nbs1) complex. Once at the break, ATM that becomes activated, phosphorylating multiple substrates. In a reaction that depends in multiple endo and exonuclease activities (including Mre11, Exo1 and CtIP) DSBs are resected forming ssDNA strands. These ssDNA regions attract Rad51 and other associated proteins. The Rad51-coated nucleoprotein filaments then invade the undamaged sister strands forming HJ structures. HR is completed by new DNA synthesis and still to be identified HJ “resolvase” enzymes.;https://www.intechopen.com/
Radiation Damage:
Radiations like Gamma and X-rays can also cause ionizations or ring opening in bases, or they can open sugar rings or break strands and even they can cause chromosomal breaks. The radiations that cause ionization of bases can lead to wrong base pairing. This is one of the methods used to generate large number of mutations in plant breeding to generate new strains or varieties.
- On the other hand UV radiations at 254nm wave length can easily cause, if the exposure is direct and sufficient in quantity and duration, cross linking between two neighboring Thymidine bases to form Thymidine-Thymidine dimers by 5’-5’ linkage and 6’-6’linkage or Thymidine-cytosine dimer by 6’-6’ linkages to produce cyclobutane rings in the same strand. An even cytosine -cytosine linkage as dimer is possible. The deadly dimerization is possible between G and G of opposite strands.
A daily dosage of daylight UV exposure, skin cells can accumulate 1000 nucleotides of Thymidine dimers per day. Dimrization prevents normal base pairing and disrupts continuous new strand formation during replication. This leads to skin cancer, which is called Xeroderma pigmentosa. Some people who work daily in very hot sun light are prone to such diseases than others.
- The said disease is due to defective gene for Photolyase.
In some cases the linking can be inter strands, which has devastating effects.
If radiations cause ionization in bases, that can lead to wrong base pairing in subsequent replication.

DNA interstrand crosslink repair and cancer Interstrand cross linking between G and G

Thymidine dimers C5-C5 and C6-C6 linkage; http://mb207.blogspot.com

Thymine dimerization induced by UV light ;
http://yxsj.baiduyy.com
- If 1 rad of X-rays combines with small doses of UV radiations can be deadly and it is equivalent to 100 rads of X-ray dosage.
Highly reactive free radical induced damage:
Hyperbaric oxygen, gamma and UV radiations, ozone, peroxides and recycling drugs can cause serious damage to tissues; all of them adversely affect the tissues. Reactive oxygen radicals can cause severe damages via H2O2 production. However the activation superoxide dismutase (SOD) can take care of it, but if free radicals are produced in large amounts and for longer duration the effect will be devastating
O2---->O2^- -- (super oxide) --->H2O2---> OH^- (OH radical) ---> H2O
Cells contain a variety of metal ions and many a times they generate free radicals such as iron and other metal ions. These ions are highly reactive and they can cause damage to DNA in the form of strand break or modifying bases or opening of rings. Free radicals are energy rich ions and they can easily attack deoxy ribose sugars and break the ring structure to generate opened rings with 3’phosphate groups. These free radicals also cause tissue damage. Such radicals generate during blood transfusion and also during certain biochemical reactions, where hydrogen peroxide generating enzymes are involved in the reactions. The damage to DNA by oxygen radicals is mediated by metal ions especially Fe^2+ ions, which leads to Fenton reactions.
Fe^2+ + H2O2 + H^+ -----> Fe^3+ + ^-OH + H2O
Cells do have enzyme to combat intracellular free radical damage. One such enzyme is super oxide dismutases (SOD), which acts like scavengers of free radicals and remove them from circulation. Among many free radicals, H2O2^- very reactive and they can act on deoxyribose sugar rings and break them to generate 3’P-open sugar rings. Even iron free radicals can severely damage DNA.
Heat and Acid induced damages:
Within the body of an organism, most of the tissues are subjected to changes in pH and body temperature. Eating and drinking of hot food or liquid can cause damages to gut, stomach and intestinal tracks; they are the most abused systems. This can cause the loss of bases, because glycoside bonds between the base and the sugar residues for they are very week. When glycosidic bonds break, the bases can easily break, thus create apurinic sites. In the region of apurinic sites the sugar-phosphate-sugar backbone remains intact. In most of the cases guanine is removed and infrequently adenine is also removed. However it is not surprising to observe depyriminidation. In all these cases holes are generated. A single genome can suffer about 10000 base losses per day. If these depurination and depyriminidation are not corrected, cells don’t work even for a single day.
Chemical induced damages:
Ames test:
Ames test is one of the standard test to identify a chemical or an agent that can induce mutations which can be identified easily. Chemical damages to DNA can have devastating effect on environment and organisms. Chemical induced mutation can be detected by a famous test called Ames test.
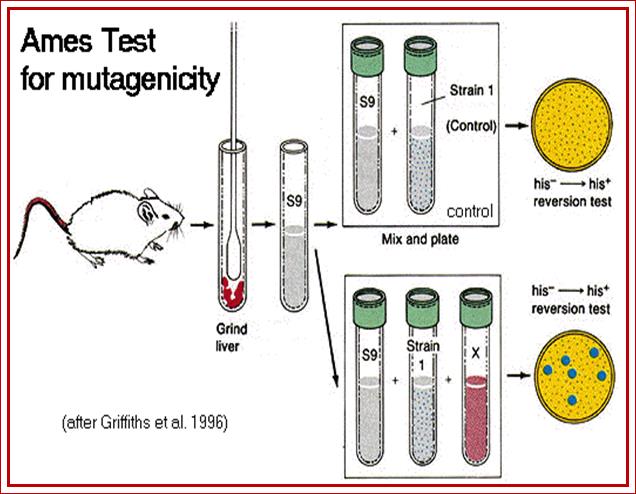
The Ames Test for mammalian environmental mutagenicity; http://www.mun.ca/
Ames devised a simple but an effective test to detect mutagenic effect of chemicals on Salmonella typhimurium strains that are defective in Histidine pathway. When cells are grown on a medium without histidine as a nutrient or metabolite supplement the cell fail to grow. When such cells treated with chemical mutagens to be tested and then plated on to minimal medium, only those mutants that repaired the gene to normal Histidine synthesis grow, others won’t. This gives an identity that the said chemical is capable of causing mutation. Then this strain can be used for further investigation, where one can study, what kind of mutation has occurred.

Ames test and detection of mutation; http://www.mun.ca/
Studies have shown that a whole lot of chemicals cause damage to DNA in one form or the other. The list of chemicals that can cause damage is a long one; here only some important ones are quoted.
- Hydroxyl amine-cause amination
- Dimethyl sulphate or dimethyl-sulfoxide-add methyl group.
- N’methyl N’nitro-n-nitrosoguanidine- adds methyl group
- 2’Amino purine- analogue base pairs with T
- 2,6’ Di amino purine- analogue
- 5’Bromodeoxyuracil- base anologue base pairs with Guanine.
- Nitrous acid-remove NH2 groups.
- Ethyl methane Sulphate-adds methyl group
- Ethyl ethane Sulphate-add ethyl group.
- Proflavins-distort ds DNA by interaction.
- Acridine orange-distort ds DNA by intercalating among the base pair stacks.
- Ethidium bromide-distort dsDNA by interaction, by substitution, change reading frame.
- Methotrexate-inhibit DHFR protein
- Aminopterin-inhibit Thymidine biosynthesis.
- 5’Fluorodeoxyuridine.
- 6’Mercaptopurine.
- Benzoyl (a) pyrene.
- Dialkylbenzanthracene.
- Anthracene.
- Dialkyl benzylanthracene.
- Afflotoxins- bind to bases and produce very heavy adducts.
- Methyl cholanthrene.
- Phorbal esters
- Mitomycin-cross link opposite strands of DNA.
- Bleomycin- cause break of DNA strands.
- 8’azaguanine- incorporated in place of G, pair with A, U, C.
- 6’Azauracil-anlogue.
- 5’Fluorouracil
- Thioguanine-analogue.
- Hydrazine
- Hydroxyuracil
- Thioguanine
- Mercaptopurine
- Diamethane
- MnCl2
- Dimethyl nitrosamine
- Sodium bisulfite- causes deamination.
- Dimethyl sulfoxide-alkylate G at 7 position
Deamination reactions:
Nitrous acid causes Deamination of cytosine, adenine, guanine and 5’methyl cytosine.
- Nitrous acid treatment and subsequent oxidation converts cytosine into Uracil. Then Uracil base pairs with adenine. Uracil DNA glycosylase enzymes remove most of the deaminated cytosines.
If 5- methylated cytosines are deaminated, they produce thymidine and the glycosylase enzyme does not act on Thymidines; this results in a change in the sequence and cause mutations (transition mutations). Such 5-methylated cytosines sites act as mutation hot spots. Lac-I genes in bacteria one finds at least 80 mutational hot spots and this is due to the presence of 5-methylated cytosine sites.

How deamination reactions change the character of bases. http://www.sivabio.50webs.com/
Cytosine deamination reactions

https://biotechkhan.wordpress.com
Bases in DNA can change other forms, which change the codon information
Cytosine + HNO2 + (O) -----> Uracil,
In G=C base pairing if C-->converted to Uracil*, in the next round of replication U* base pairs with A (U*= A), in the next round of replication, in the place of U, Thymidine will be incorporated,
U = A----> T = A,
Here the C = G base pair is converted to T =A.
- Adenine + HNO2 + (O) ------> Hypoxanthin,
Hypoxanthin base pairs with cytosine,
A = T------> Hypoxanthine = T -----> HPX = C ------> G = C.
A=T base pairs are changed to G=C base pairing.
- Similarly Guanine on treatment with Nitrous acid and subsequent oxidation results in the formation of Xanthine, which base pairs with cytosine.
G* = C à Xanthine*, ----> X = C, ------> C =G.
In this G=C is changed to C=G.
- On nitrous acid treatment 5-methyl cytosine is converted to Thymidine, which pairs with Adenine,
5’CH3-C = G-----> T = G, ---T = A, C=G is converted to T = A. The bases thus produced are not removed and remain so. Thus these regions containing 5’methyl cytosines considered as mutational hot spots.
Change of 5’CH3 C=G to A=T
- Sodium bisulfate also causes deamination, where Cytosine is converted to Uracil.
C = G ----> C is converted to U, then U pairs with A, the consequence is C=G is converted to U = A and U=A is converted T=A
- Spontaneous deamination cum oxidation of cytosine results in Uracil and this happens at the rate of 100 bases per genome per day.
· Spontaneous deamination of cytosine is also a process by means of
which 100 or more deamination reactions take place per genome per day?
Amination reactions:
Hydroxylamine treatment causes cytosine to be aminated at 6th position to produce 6- Hydroxylamine cytosine. This hydroxylamine group is a heavy adduct, which is not desirable.
Base analogues:
There are many base analogues, which can be incorporated into DNA as substitutes. When such analogues get incorporated, they change base pairing subsequently, thus cause change in the information.
- 5=Bromo deoxy Uracil (BURD) is a substitute for Thymidine when incorporated pairs with Adenine. The ionized BURD base pairs with Guanine.
BURD pairs with Guanine, A=T is changed to A=BuRD, BuRD=G,
So A=T is changed to G=C
Similarly 2’aminopurine and 2, 6’ di aminopurine can act as substitute and provide base pairing. The 2’ aminopurine when incorporated pairs with Thymidine. But if this is ionized, it can easily base pair with cytosine.
- NH2 purine = Thymidine,
- NH2 purine = Cytosine,
Such base analogues are used in vitro mutational studies; they are 8’Azaguanine, 6’Azauracil, 5’ Fluorouracil and Thioguanine. Azaguanine is used in place of Guanine for sequencing DNA, which is rich in GC contents. Ionosine is another base, which can be incorporated as a nucleotide, and this nucleotide can pair with Adenine, Uracil and Cytosine.
Ionized Bases and Oxidative Bases:
As mentioned above ionized bases substitute base pair differently. Ionized Thymidine, Adenine and Cytosine, instead of base pairing with Adenine, Thymidine and Guanine, they base pair with Guanine, Guanine and Guanine (the last one with Hoogsteen base pairing) respectively.
T = A, ionized T instead base pairs with guanine; T*= G; -> C=G
A = T, ionized A base pairs with Guanine; A* = G;-> C=G
C = G, ionized C pairs with guanine by Hoogsteen mode, C* = G.
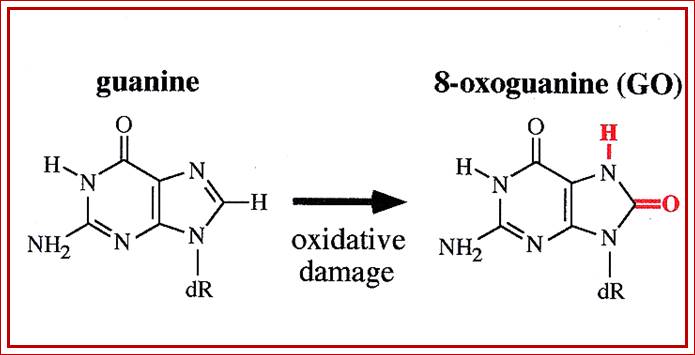
Guanine can be oxidized to 8-oxoguanine; https://en.wikipedia.org

O-6 methylation of Guanine base pairs; http://www.atdbio.com
Alkylating agents:
There are several Alkylating chemicals, which transfer either ethyl or methyl groups to reacting positions of the bases, which can lead to hydrolysis of glycoside bond between the base and the sugar. This causes Apurinic or Apyrimidinic sites, leaving phosphate-sugar-phosphate backbone intact.
- Dimethyl sulphoxide methylates Guanine with great avidity to produce 7-methyl Guanine. If such 7’methylated guanine containing DNA is treated with piperidine, the DNA breaks the S-p-S backbone before the Guanine nucleotide. It cleaves at Guanine more frequently than cleaving at Adenine nucleotide. If the guanine methylated DNA is treated with Formic acid, the cleavage is specific at Guanine.
DNA + Dimethyl sulphoxide--> 7’CH3-G DNA.
7’CH3-G.DNA + Piperidine -> 5’ (p-G-DNA) n (more frequently) + 5’ (p-A-DNA) rarely.
7’CH3-G-DNA + Formic acid -> 5’ (P-G-DNA) n.
If the DNA is treated with Hydrazine followed by piperidine, the cleavage is before Cytidine and Thymidine, but if the DNA is treated with hydrazine in the presence of 1.5mM NaCl, cleavage is preferred only at Cytidine.
- Alkylating agents can also methylate guanine at 6’- O position. If methylated guanine is not removed, it can pair with thymidine and subsequent replication changes the information.
C=Gà C= CH3-G, leads to CH3G pairs with Thymidine, and then Thymidine pairs with Adenine.
CH3G=T à A=T
Most of the alkylating agents transfer their reactive methyl groups to suitable bases at specific positions. This kind of alkylation, generally leads to the break down of the glycosidic bonds and also breakdown of p-s-p backbone at depurinated sites.
- N-methyl N-Nitro N’nitrosoguanidine is a powerful alkylating agent and most preferably act at replication forks and change the base sequences in clusters. N’methyl-N’Nitro-n-Nitrosoguanidine transfers its methyl group to guanine to produce 6’O-guanine.
Ethyl methane sulphonate transfers ethyl group to Guanine to its 6’O position creating 6’O-CH2 guanidine. This kind of ethylation can facilitate the O-ethylated Guanosine to pair with thymidine.
C=Gà C=CH2.Gà T=CH2.Gà T=A
- Di-methyl nitrosamine and di-ethyl nitrosamine have very reactive methyl and ethyl groups and such groups can be easily transferred to the receiving positions provided the enzymes are in place. Cells do have specific enzymes, which methylate bases at specific positions in sequence dependent manner. Cytosine methylases methylate cytosine at 5’ position. This methylation is sequence dependent and the sequence is CGCGCG. The deoxy cytosine methylases use S-Adenosyl homo-cysteine, which is the donor of methyl group. Another methylase, S-Adenosyl methylase, methylates adenine at N6’ position only when the sequence is GATC and the enzyme uses S-Adenosyl methionine as the substrate. The SAM methylase can also methylate Guanine nucleotide, but rarely and under certain controlled condition. Not just bases, even phosphates of P-S-P backbone can also be methylated.
If E.coli cells are exposed to higher levels of nitroso-guanine or to any other alkylating compounds, cells become resistant to mutagenic and toxic effects of mutagens. This is because, the drugs induce the synthesis of O-6-methyl guanine by methyl transferase, which is responsible for the removal of methyl group from O-6 th position, at the same time another enzyme that is induced is Glycosylase, which removes the modified bases and creates Apurinic or Apyrimidinic sites. The induction is 100 fold and thus cells become resistant to the methylating drugs. A group of genes called ‘Ada’ produce these enzymes, and these genes are positively regulated.
Effect of Bleomycin: It is an antibiotic and it can cause strand breaks.
Mitomycin effect: It is another antibiotic, which react with ds DNA strands and brings about covalent linkages between complementary bases. This distorts the structure of the DNA prevents the separation of ds DNA at the time of replication. It can also cause strand breaks.
Effect of Acridine, Proflavins and Ethidium dyes:
These dyes are fluorescent compounds, they intercalate or insert the dyes into spaces found in between flat base pairs and lengthen the DNA. Such lengthening of the DNA can cause either deletion or addition during next round of replication, these are manifested as bulges.
Effect of chemical carcinogens: Afflotoxin, Benzo (a) pyrene and Methyl cholanthrene:

https://yourhealthofima.wordpress.com
Afflotoxin are the products of Fungus Amanita species, Benzo (a) pyrene is the product of burnt food or burnt gasoline found in emissions. They bind and covalently link to bases as bulky adducts. Afflotoxin by alkylation at N7th position can lead to the break of P-S-P bonds. Potato chips and French fries fried in oil produce acrylic compounds, which are carcinogenic.

Benzo(a)pyrene, a chemical produced by internal combusion engines and thus common in the environment, is not itself mutagenic. However, in the mammalian liver, benzo(a)pyrene is metabolized to diol epoxide, which binds covalently to guanine bases, preventing proper base pairing with cytosine bases. Bulky Addition Products such as diol epoxide or Aflatoxin B1 may result in depurination mutagenesis and are known to be carcinogens.;http://www.mun.ca




Carcinogen bound foodmaterials; https://aghealth.wordpress.com;https://www.daf.qld.gov.au;http://www.yourhealthremedy.com http://bioweb.uwlax.edu

https://www.chegg.com
Refined solution structure of 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 opposite CpA in the complementary strand of an oligodeoxynucleotide duplex as determined by 1H NMR.
Johnston, D.S., Stone, M.P.Afflotoxins binds to DNA as adduct; http://www.rcsb.org
(a)
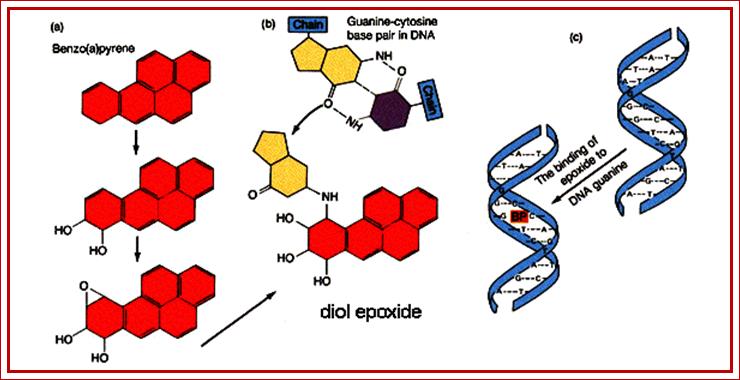
Benzo(a)pyrene, a chemical produced by internal combusion engines and thus common in the environment, is not itself mutagenic. However, in the mammalian liver, benzo(a)pyrene is metabolized to diol epoxide, which binds covalently to guanine bases, preventing proper base pairing with cytosine bases. Bulky Addition Products such as diol epoxide or Aflatoxin B1 may result in depurination mutagenesis and are known to be carcinogens; inding of carcinogen leads to change in Guanine affinity to cytosine; http://www.mun.ca
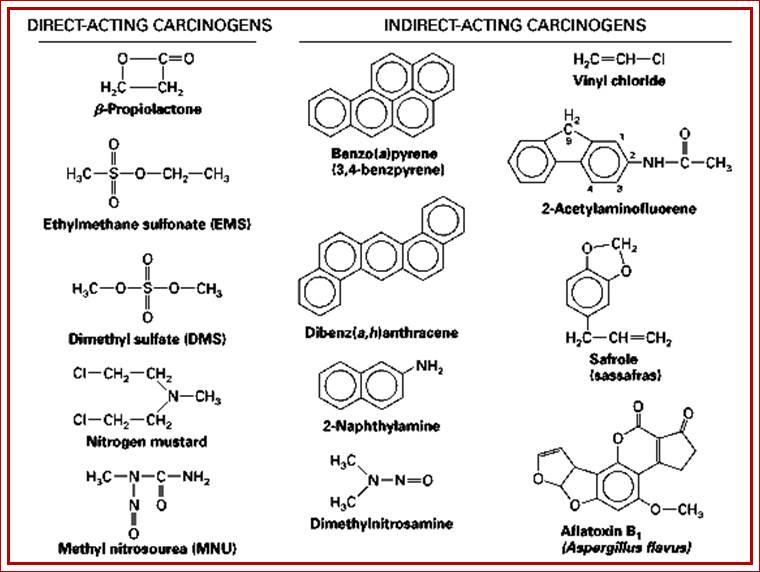
Some carcinogens; http://www.biologydiscussion.com
From the above account it can be understood why DNA is damaged and how DNA can be damaged. The following are few important forms of damages:
- Missing base: Removal of purines by acid and heat. This happens in every cell, in every genome, at the rate of 10^4 per day per genome. Deamination of cytosine to Uracil and removal of Uracil by glycosylases on large scale is an every day event.
- Mismatched base pairing: Replication though accurate and shows high fidelity, wrong nucleotides are incorporated, this in spite of the activity of molecular proof reading enzymes, some mismatch errors persists at post replication stage. An increased rate of incorporation of wrong bases happens when genes responsible for proof reading processes, like epsilon subunit of DNA pol-III or DNA pol-I itself suffer with mutations. This also true for Eukaryotes where enzyme subunits involved in 3’-5’ exonuclease proof reading subunits. Even some defective nuclease can also create such errors.
- Altered bases: Ionization of bases, tuatomeric, base analogues, amination, deamination and alkylations modify bases and they in turn base pair differently and cause damage to DNA in terms of information.
- Dimrization bases by cross linking: UV radiations at 260-310-nm can induce breakage of double bonds in heterocyclic rings of bases and activate bases located at neighborhood or side by side to cross link to form T-T dimers or T-C dimers.
- Inter-strand cross-linking: Mitomycin can cause inter-strand cross-linking.
- Addition and deletion bulges: Acridine and Proflavins intercalate into spaces found between stacked base pairs, extend the length of the DNA and cause deletions and additions and thus produce bulge.
- Ring openings, strand breaks: High-energy free radicals in association with certain metal ions can cause break in the deoxyribose rings to generate strand breaks with 3’phophate ends. Radiations like X-rays and Gamma rays can easily break DNA strands.
Combination of X-rays and UV radiations is deadly.
- Addition of heavy adducts: Afflotoxin, methyl cholanthrene and such compounds are added to bases causing heavy adducts and distort DNA structure.
In all the above said cases, cells are endowed with factors and enzymes to sense the kind of damage, respond to different types of damages and repair the DNA before it is perpetuated. In spite of this kind of molecular vigilance some damages escape the repairs system and cause heritable defect, which can be devastating, or cancer in a lifetime of an individual.
Types and frequency of DNA damage on daily basis:
|
Damage |
Events/day |
% frequency/day |
|
Single strand break |
120,000 |
50.9 |
|
N^7CH3guanine |
84,000 |
35.6 |
|
Depurination |
24,000 |
10.2 |
|
O^6CH3guanine |
3120 |
1.3 |
|
Oxidized DNA |
2880 |
1.2 |
|
Depyrimidination |
1320 |
0.5 |
|
Demaination-cytosie |
360 |
0.2 |
|
Double strand braks |
9 |
0.001 |
|
Interstrand cross link/dimerization |
8 |
0.01 |
|
|
|
|
CONTENTS
1. 1Introduction
2. 2DNA damage
1. 2.1Mismatches in DNA bases
1. 2.1.1The G·T mismatch
2. 2.1.2The A·C mismatch
3. 2.1.3Minor tautomer mismatches
4. 2.1.4The G·A mismatch
5. 2.1.5Inosine-containing mismatches
6. 2.1.6Measuring the stability of mismatch base pairs using UV melting
7. 2.1.7Shape and stability of base pairs
2. 2.2Chemical damage to DNA bases
1. 2.2.1Deamination of cytosine
2. 2.2.2Methylation of guanine
3. 2.2.3Oxidation of adenine and guanine
1. 3.3Direct reversal
2. 3.4Base excision repair
3. 3.5Nucleotide excision repair
4. 3.6Mismatch repair
1. 3.6.1Distinguishing parent and daughter DNA strands: DNA methylation
2. 3.6.2Mechanism of mismatch repair
5. 3.7Other DNA repair mechanisms
4. 4When damaged DNA is not repaired
1. 4.1Evolution