DNA C- Value paradox
1. Introduction:
Quantity of DNA in an organism per cell, in all cells, is always constant, for a given species. Given the list of organisms on this planet, with teaming millions, each have its own genome whose size varies from one species to the other and no two species have the same amount of genome nor the same genomic value or character? At this point of time, nobody has found two different species, however close in phylogeny they may be, have the same size of the genome, even one finds such situations, and some of their sequence will be different. However when haploid genomic content is quantitated species wise, phylum wise, or kingdom wise in increased order of evolutionary table, one finds enormous variation, not only in the same phylum, group, order, family or genus or species. Variation in genomic content (both qualitatively and quantitatively) within a phylum or an order or genus is surprisingly large from 10^5 bp to 10^12 bps. Animals show variations range more than 3,300-fold, and in land plants among them differ by a factor of about 1,000. Protists genomes have been surprisingly vary more than 300,000-fold in size.
Origin of the term the term C:
“What is C Value- is it the Content, Complement, Concentration or what?, if it is content then what content, 2n or n. Many authors have assumed that the "C" in "C-value" refers to "characteristic", "content", or "complement". Even among authors who have attempted to trace the origin of the term, there had been some confusion because Hewson Swift did not define it explicitly when he coined the term C’ value in 1950. In his original paper, Swift appeared to use the designation "1C value", "2C value", etc., in reference to "classes" of DNA content (e.g., Gregory 2001, 2002); however, Swift explained in personal correspondence to Prof. Michael D. Bennett in 1975 that "I am afraid that the letter C’ stood for nothing more glamorous than 'constant', i.e., the amount of DNA that was characteristic of a particular genotype" (quoted in Bennett and Leitch 2005). This is in reference to the report in 1948 by Vendrely and Vendrely of a "remarkable constancy in the nuclear DNA content of all the cells in all the individuals within a given animal species" (translated from the original French). Swift's study of this topic related specifically to variation (or lack thereof) among chromosome sets in different cell types within individuals, but his notation evolved into "C-value" in reference to the haploid DNA content of individual species and retains this usage today”.
However the discovery of a large amount of non-coding DNA lead to the concept of C-DNA value or C-Value paradox and variation is surprisingly so vast it is called C-DNA value paradox. The paradox or the enigma is between the C-value and the gene numbers. Elucidation of noncoding DNA and noncoding but functional RNA can resolve this. The disjunction between the human genome complexity-number of genes in organism and such complexity is termed as G-value paradox; distinguish between C-value paradox and G-value paradox? The information value the I-value is about the information found in the G value complex.
|
Species |
C-value, x 10^6 |
G value, no. of genes |
I-value-Information on genes |
|
S.cerevisiae |
12 |
6000 |
|
|
D. melanogaster |
130 |
14000 |
|
|
C.elegans |
97 |
19000 |
|
|
A.thaliana |
125 |
26000 |
|
|
H.sapiens |
2900 |
31000(2002), ~21000 |
|
|
|
|
|
|
|
|
|
|
|
C Value: The amount DNA found in haploid genome, measured in million base pairs or in pg; the C may mean constancy of the genome in the species.
G Value: The number of genes found in the haploid genome; the number includes predicted and known ORFs.
I value: The amount of information embedded in the genome; their estimates effective number of genes which encompasses alternative splicing, post translational modifications, multi-domain proteins and gene redundancy plus gene expression and gene interaction.
Comparison of Different Genome Sizes
Genome sizes:
Since genomes are very complex, one research strategy is to reduce the number of genes in a genome to the bare minimum and still have the organism in question survive. There is experimental work being done on minimal genomes for single cell organisms as well as minimal genomes for multi-cellular organisms (seeDevelopmental biology). The work is both in vivo and in silico.
|
Organism type |
Organism |
Genome
size |
Note |
|
|
Porcine circovirus type 1 |
1,759 |
1.8kb |
Smallest viruses replicating autonomously ineukaryotic cells.[19] |
|
|
3,569 |
3.5kb |
First sequenced RNA-genome[20] |
||
|
5,224 |
5.2kb |
|||
|
5,386 |
5.4kb |
First sequenced DNA-genome[22] |
||
|
9,749 |
9.7kb |
|||
|
48,502 |
48kb |
Often used as a vector for the cloning of recombinant DNA. |
||
|
1,259,197 |
1.3Mb |
Until 2013 the largest known viral genome.[27] |
||
|
2,470,000 |
2.47Mb |
Largest known viral genome.[28] |
||
|
1,830,000 |
1.8Mb |
First genome of a living organism sequenced, July 1995[29] |
||
|
Nasuia deltocephalinicola(strain NAS-ALF) |
112,091 |
112kb |
Smallest non-viral genome.[30] |
|
|
159,662 |
160kb |
|||
|
600,000 |
600kb |
|||
|
700,000 |
700Kb |
|||
|
4,600,000 |
4.6Mb |
|||
|
Solibacter usitatus (strain Ellin 6076) |
9,970,000 |
10Mb |
||
|
Polychaos dubium ("Amoeba" dubia) |
670,000,000,000 |
670Gb |
||
|
157,000,000 |
157Mb |
First plant genome sequenced, December 2000.[36] |
||
|
63,400,000 |
63Mb |
Smallest recorded flowering plant genome, 2006.[36] |
||
|
Fritillaria assyrica |
130,000,000,000 |
130Gb |
||
|
480,000,000 |
480Mb |
First tree genome sequenced, September 2006[37] |
||
|
Paris japonica (Japanese-native, pale-petal) |
150,000,000,000 |
150Gb |
Largest plant genome known |
|
|
480,000,000 |
480Mb |
|||
|
12,100,000 |
12.1Mb |
First eukaryotic genome sequenced, 1996[39] |
||
|
30,000,000 |
30Mb |
|||
|
100,300,000 |
100Mb |
First multicellular animal genome sequenced, December 1998[40] |
||
|
20,000,000 |
20Mb |
Smallest animal genome known[41] |
||
|
Drosophila melanogaster (fruit fly) |
130,000,000 |
130Mb |
||
|
Bombyx mori (silk moth) |
432,000,000 |
432Mb |
14,623 predicted genes[43] |
|
|
Apis mellifera (honey bee) |
236,000,000 |
236Mb |
||
|
Solenopsis invicta (fire ant) |
480,000,000 |
480Mb |
||
|
Tetraodon nigroviridis (type of puffer fish) |
385,000,000 |
390Mb |
Smallest vertebrate genome known estimated to be 340 Mb[45][46] - 385 Mb.[47] |
|
|
2,700,000,000 |
2.7Gb |
|||
|
3,200,000,000 |
3.2Gb |
Homo sapiens estimated genome size 3.2 billion bp[49] Initial sequencing and analysis of the human genome[50] |
||
|
Protopterus aethiopicus(marbled lungfish) |
130,000,000,000 |
130Gb |
Largest vertebrate genome known |
|
WIKIPEDIA
Proportion of non-repetitive DNA;
Since genomes and their organisms are very complex, one research strategy is to reduce the number of genes in a genome to the bare minimum and still have the organism in question work. There is an experimental work being done on minimal genomes for single cell organisms as well as minimal genomes for multicellular organisms (see Developmental biology). The work is both in vivo and in silico. WIKIPEDIA
“Genome sizes are typically given as gametic nuclear haploid DNA contents (‘C-values’) either in units of mass (picograms, where 1 pg = 10−12 g) or in number of base pairs , 1 Mb = 106 base pairs. These are directly interconvertible as 1 pg = 978 Mb (or 1 Mb = 1.022 × 10−3 pg)”. The quantity of the cDNA varies 8000 fold in eukaryote genomes. If human 23 chromosomal DNAs are joined end to end , it will be 2meters long and 19-10°A thick”.
Range of Genomic Content Among the groups:
Mycoplasma 10^5-6 bp
(-) Bacteria 4.2 to 5x10^6 bp
(+) Bacteria 2 to 8x10^6 bp
Fungi 2 to 5x10^7 bp
Algae 5 to 8x10^7 bp
Molds 6 to 9x10^7 bp,
Worms 7x10^7 to 2x10^8 bp,
Molluscs 6x10^8 to 7x10^9 bp,
Insects 1.5x10^8 to 6x10^9 bp,
Echinoderms 5x10^8 to 5x10^9 bp,
Cartilage fishes 3x10^9 to8x10^9 bp,
Bony fishes 6x10^8 to 9x10^9 bp,
Amphibians 8x10^8 to 9x10^11 bp,
Reptiles 2x10^9 to 5x10^9 bp,
Birds 2x10^9 to 9x10^9 bp,
Mammals 3x10^9 to 5x10^9 bp,
Flowering plants and few animals-Range from 8 x 10^8 to 2 x 10^12 bp
Frittilaria assyriaca.132pgx978Mbp =1.2X10^5Mbp =1.2x10^11bp.
Pieris (Paris) japonica. 152pgx978Mbps =1.486x10^11 bp.
Psilotum nudum. 2.5 x 10^11 bp.
Protopterus aethiopicus, animal, 132pg (132,000,000,000).
Polychaos dubium (Ameba dubia ), 670 x10^9 bp or 6.7x10^11bp.
Note- 1pg = 978 mb or million base pairs or 978x10^6bp = 0.978x10^9 bp
Genome size and cell size: The term Genome’ was coined by H. Winkler in 1920.The term C-value was coined by Swift 1950 with reference to haloid genome. The most universal effect of genome size increase is a concomitant increase in cell size (Gregory2001). While early genome size biologists were surprised to discover that genome size did not correlate with apparent organismal complexity, positive correlations between erythrocyte size and genome size have long been noted in vertebrates (Mirsky and Ris1951). Subsequent studies have confirmed this relationship between genome size and cell size in Protists (Cavalier-Smith1985), plants (Gregory2005a ), invertebrates (Gregory2005b), amphibians(Olmo and Morescalchi1975; Horner and Macgregor 1983), reptiles (Olmo and Odierna 1982), fish(Pedersen1971; Olmo1983; Hardie and Hebert 2003), birds (Gregory2002a ), and mammals (Gregory2000). Numerous theories have been invoked to account for the strong correlation between genome size and cell size (Gregory 2005a). Among these, the nucleo-skeletal theory states that cell size is adjusted adaptively in response to selective pressures, and these changes enjoin correlated shifts in nucleus size (Cavalier-Smith1985). Comparison of haploids (rare in plants) with diploids and polyploids show some interesting features such as size of plants and variability.
A guided tour of large genome size in animals what we know and where we are heading. http”//academia.edu/1062582
How Many Protein-Coding Genes are in a Genome? Interestingly, the same "remarkable lack of correspondence" can be noted when discussing the relationship between the number of protein-coding genes and organism complexity. Scientists estimate that the human genome, for example, has about 20,000 to 25,000 protein-coding genes. Before completion of the draft sequence of the Human Genome Project in 2001, scientists made bets as to how many genes were in the human genome. Most predictions were between about 30,000 and 100,000. Nobody expected a figure as low as ~21,000, especially when compared to the number of protein-coding genes in an organism like Trichomonas vaginalis. T. vaginalis is a single-celled parasitic organism responsible for an estimated 180 million urogenital tract infections in humans every year. This tiny organism features the largest number of protein-coding genes of any eukaryotic genome sequenced to date: approximately 60,000 (?).
In fact, compared to almost any other organism, humans' ~20-25,000 protein-coding genes do not seem like many. The fruit fly Drosophila melanogaster, for example, has an estimated 13,000 protein-coding genes. Or consider the mustard plant Arabidopsis thaliana, the "fruit fly" of the plant world, which scientists use as a model organism for studying plant molecular genetics. A. thaliana has just about the same number of protein-coding genes as humans—actually, it has slightly more, coming in at about 25,500. Moreover, A. thaliana has one of the smallest genomes in the plant world! It would seem obvious that humans would have more protein-coding genes than plants, but that is not the case. These observations suggest that there is more to the genome than protein-coding genes alone.
As shown in Table 1 (adapted from Van Straalen & Roelofs, 2006), there is no clear correspondence between genome size and number of protein-coding genes—another indication that the number of genes in a eukaryotic genome reveals little about organismal complexity. The number of protein-coding genes usually caps off at around 25,000 or so, even as genome size increases. If one calculates a gene that codes for a protein of 300 amino acids, it means the ORF of the gene is 300x3 = 900 bp. Add another 150-200bp for 5UTR and 3’UTR, it will be 1100bp. But the gene contains non coding introns, whose size and number varies, thus it is difficult calculate the size of the genome based on protein coding part of the genes for there are other regions of DNA which does not code for proteins, but code for functional RNAs, an some don’t code for anything.
Table 1: Genome Size and Number of Protein-Coding Genes for a Select Handful of Species
|
Species and Common Name |
Estimated Total Size of Genome (bp)* |
Estimated Number of Protein-Encoding Genes* |
|
Saccharomyces cerevisiae (unicellular budding yeast) |
12 million |
6,000 |
|
Trichomonas vaginalis |
160 million |
60,000 |
|
Plasmodium falciparum (unicellular malaria parasite) |
23 million |
5,000 |
|
Caenorhabditis elegans (nematode) |
95.5 million |
18,000 |
|
Drosophila melanogaster (fruit fly) |
170 million |
14,000 |
|
Arabidopsis thaliana (mustard; thale cress) |
125 million |
25,000 |
|
Oryza sativa (rice) |
470 million |
51,000 |
|
Gallus gallus (chicken) |
1 billion |
20,000-23,000 |
|
Canis familiaris (domestic dog) |
2.4 billion |
19,000 |
|
Mus musculus (laboratory mouse) |
2.5 billion |
30,000 |
|
Homo sapiens (human) |
2.9 billion |
20,000-25,000 |
* There may be other estimates in the literature, but most estimates approximate those listed here. Eukaryotic Genome Complexity By: Leslie A. Pray, Ph.D. © 2008 Nature Education
Paradox:
If one scans organisms from simplest unicellular to the most complex organisms, as that of human species and some flowering plants, one finds a size difference ranging from 10^5 to 10^12 bp (plants). Consider a simple amphibian and a mammal i.e. human being, the size of the genome in both is almost the same ie.10^9 bp. Elephant Loxodonta africana africana (Elephant) genome size is 3.3 x10^9bp. But Woolly Mamoth extinct (45,000 years back) elephant genome estimated as >4x10^9bp. Take for example another mammoth , can live 120-150years, weigh 40-50 tons, the whale contains ~3x10^9 bp.
Again, no significant correlation exists between the number of genes or chromosomes and evolutionary advantage. The Balsam Poplar tree has a genome size of 0.55 Gb, 19 chromosomes, and 45,555 genes. The Carp fish has a genome size of 1.7 Gb, 100 chromosomes, and 7,127 genes (Sheema Ali). A cloned cat shows variation in its offspring, these due to epigenetic changes. Another example makes it one wonder, why there is so much of difference between a fruit fly (1.4x10^8 bp) and a house fly (8x10^8bp), where the body pattern is more or less the same with the exception of body size. If one compares the genome size of an amphibian X. laevis (10^9bp) and a lungfish (10^11bp), the difference is huge. There are many such anomalies and paradoxes difficult to explain and elucidate. Among plants Arabidopsis thaliana, an angiosperm (Dicot) known to contain smallest genome with 5 haploid chromosomes, but the first land plant called Psilotum nudum, the first vascular land plant (without roots) surprisingly consists of 2.5X 10^11 bp 3000 times the amount of Arabidopsis thaliana. The reason is due to the finding that the Psilotum contains more than 80% of its genome as what is now considered as repetitive DNA. Similarly some amphibians and lung fishes contain 30 or more amount of DNA than their counter part species. The smallest genome is found in a parasite Encephalitozoon intestinalis (a parasite in humans) contains 0.0023pg. Among plants Frittilaria assyriaca (132.52pg) (a hybrid of Trillium and Hagae) once considered to contain largest genome but Peiris japonica out-beaten it with the genome size of 152. 23 pg (1pg = 978mBP) bigger than trillium or a large marbled lung fish Proteopterus aethiopicus with 132.83pg. In comparison to them, Homo sapiens contain 3pg.
Expectedly, logic wise, as the body of an organism becomes more and more complex with more number of cells and more number of tissues and organs, the size of the genome should increase keeping in step with the complexity. But within a group, with not much of difference in the body pattern and size, function and efficiency of the organism, paradoxically one will be baffled to find large difference in the genomic content; it is very difficult to comprehend and explain why such big difference; so this puzzle is called C –Value Paradox. Study of C-DNA values of organisms, perhaps provides answer to this complexity and perplexity.
Paris japonica (152.23 pg DNA); http://rareplants.co.uk/
To explain this anomaly and the jig-saw puzzle, one has to ask a question to be answered, i.e. what is the total number of genes absolutely essential for a structure to be called life, which should exhibit growth, reproduction and live in happiest times and able to produce a population.
How many genes does it take to make an organism? Craig Venter:
“The scientists at The Institute for Genomic Research (now known as the J. Craig Venter Institute, SAN DIEGO, CA and ROCKVILLE, Lajolla) who determined the Mycoplasma genitalium genome sequence has followed this work by systematically destroying its genes (by mutating them with insertions) to see which ones are essential to life and which are dispensable. Of the 485 protein-encoding genes, they have concluded that only 381 of them are essential to life”. The species he created is named as Mycoplasma laboratorium- ‘a synthetic life’. There is speculation that this line of research could lead to producing bacteria that have been engineered to perform certain desired function. There are some fears too. It is true if such species fall into the hands of terrorists.
http://www.jcvi.org/
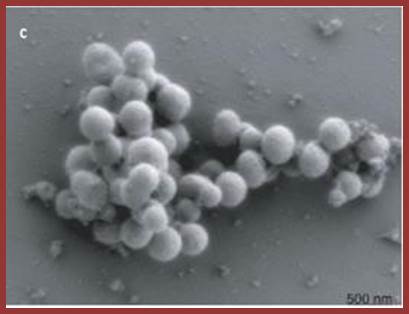
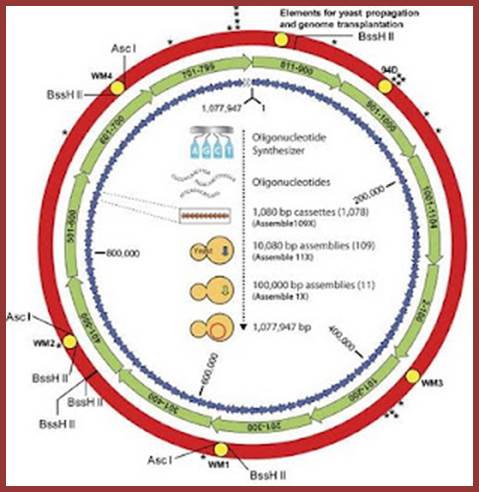
‘Craig Venter and his team have built the genome of a bacterium from scratch and incorporated it into a cell to make what they call the world's first synthetic life form’
Above is a scanning electron microscope picture of the dividing cells; Craig Venter Creates Synthetic Life form; http://www.theguardian.com/
Scientists
create First Self Replicating Synthetic Life; This is the genome that Venter
built. The text in the middle shows the process in full. The little letters
around the edge (BssH II etc) show sites for restriction enzymes which are used
to cut the genome into little pieces for analysis. Bruce Sterling; http://www.wired.com/
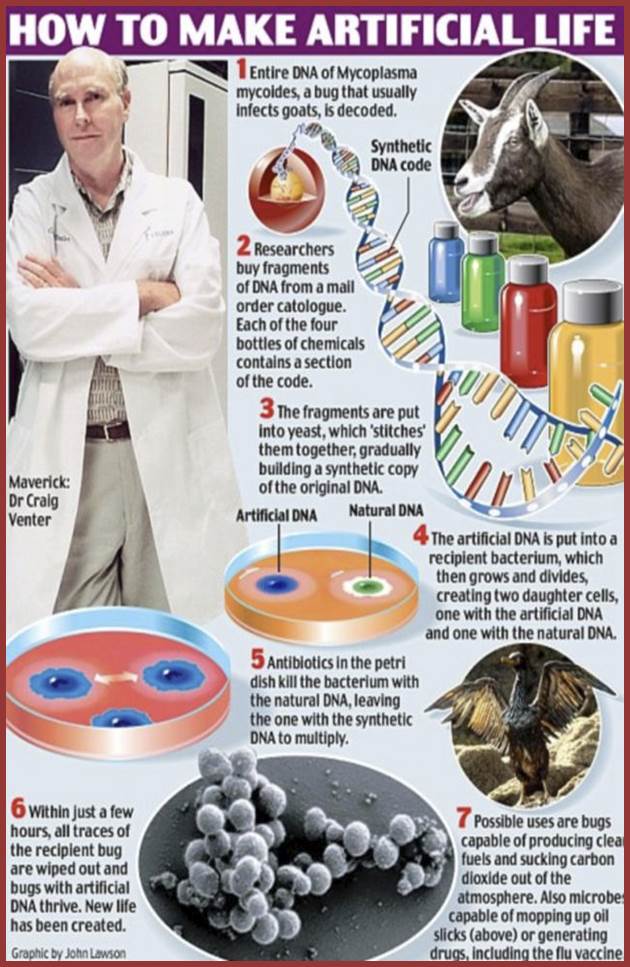
Life on Mars closer than ever for man claims scientist creating artificial life that feeds off carbon dioxide; http://www.dailymail.co.uk/
Read more: http://www.answers.com/topic/craig-venter#ixzz1sxRDMc8z. The work is remarkable. Craig Venter and James Watson are the first two people whose genome has been sequenced (PLoS Biology, and Watson's genome is available at the CSHL website).
WHO IS CRAIG VENTER? Why he was not awarded Nobel laureate?
Craig Venter is a controversial biologist and entrepreneur who led
the effort by the private sector to sequence the human genome. Frustrated with
NIH, he started ‘Celera Genomics’ funded by private company. He was passionate
with ‘shotgun library’ to complete the human genomic sequencing. After contributing to
the Human Genome, and its release into the public domain, Venter was fired by
Celera in early 2002. According to his biography, Venter was ready to leave
Celera, and but fired due to conflict with the main investor, Tony White, that
had existed since day one of the project.
Read more: http://www.answers.com/topic/craig-venter#ixzz1udaXNThV
He was vilified by the scientific community for turning the project into a competitive race but his efforts did mean that the human genome was mapped three years earlier than expected.

http://www.uncommondescent.com/
Born in 1946, Dr Venter was an average scholar with a keen interest in surfing.
It was while serving in Vietnam and tending to wounded comrades that he was inspired to become a doctor. During his medical training he excelled in research and was quick to realize the importance of decoding genes. In 1992 he set up the private Institute for Genomic Research. Then a mere three years later he stunned the scientific establishment by revealing the first complete genome of a free-living organism that causes childhood ear infections and meningitis.
In 2005 he founded the private company Synthetic Genomics, with the aim of engineering new life forms that would produce alternative fuels. He was listed on Time Magazine's 100 list of the most influential people in both 2007 and 2008.
Read more:
sciencetech/article- Artificial-life-created-Craig-Venter--wipe-humanity.htmlU. http:// www.dailymail,co.uk
DNA C value Paradox – (continued):
One can always expect, with more types of cells and complexity of cell types, more and more number of genes is required. Furthermore with the acquisition of greater number functions, regulations, adaptations and various other devises in the mode of living and reproduction, one generally expect, species should have to acquire or make greater number of genes; so the genomic size. Besides many of the DNA segments have duplicated many thousand times and added to this increase many viral mediated DNA or RNA mediated DNAs have been inserted and duplicated. More than 45% of the genomic DNA is found to be non-coding and repetitive DNA in one or the other form. From recent genomic analysis for protein coding region of the whole genome it has been found that only 1 to 1.2% of the genome accounts for it. Then the question is what is the rest of DNA doing? But recent genetic information reveals that there are more number non coding RNAs have been found.
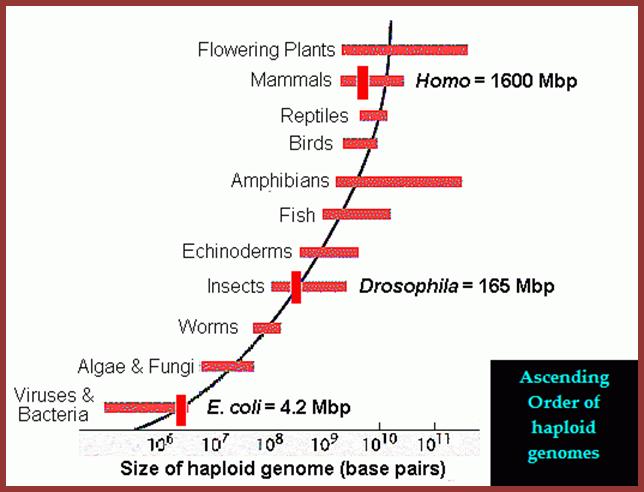 Variation n Genome Size (C-Value) among the prokaryotes
and Eukaryotes; Genome size (C-value) can be measured as
either the # bp or the mass of DNA in a haploid chromosome set, where 106 bp = 1 Mbp = 10-3 pg DNA. The actual number of base
pairs or mass of DNA in the cell of a diploid organism is therefore twice that shown here. For example, the
human euchromatic genome is 3.2
x 109 bp; http://www.mun.ca/
Variation n Genome Size (C-Value) among the prokaryotes
and Eukaryotes; Genome size (C-value) can be measured as
either the # bp or the mass of DNA in a haploid chromosome set, where 106 bp = 1 Mbp = 10-3 pg DNA. The actual number of base
pairs or mass of DNA in the cell of a diploid organism is therefore twice that shown here. For example, the
human euchromatic genome is 3.2
x 109 bp; http://www.mun.ca/
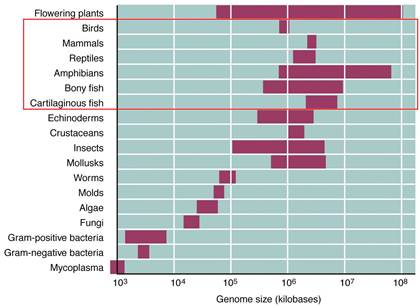
https://www.mun.ca
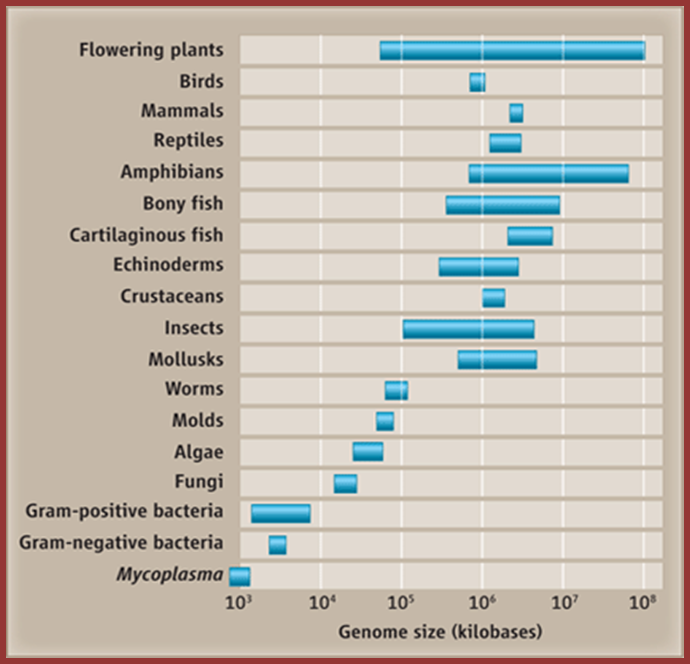
Genome sizes are not randomly distributed; Periannan Senapathy http://wasdarwinwrong.com/
This figure gives you a concept of the genome sizes from lower to higher order of organisms, of groups but looking at the size of the genome and the number genes estimated to be present in each of the genome provides you the paradox, why some of the genomes have greater amount of DNA than the required; so why such extra DNA present and what it does?
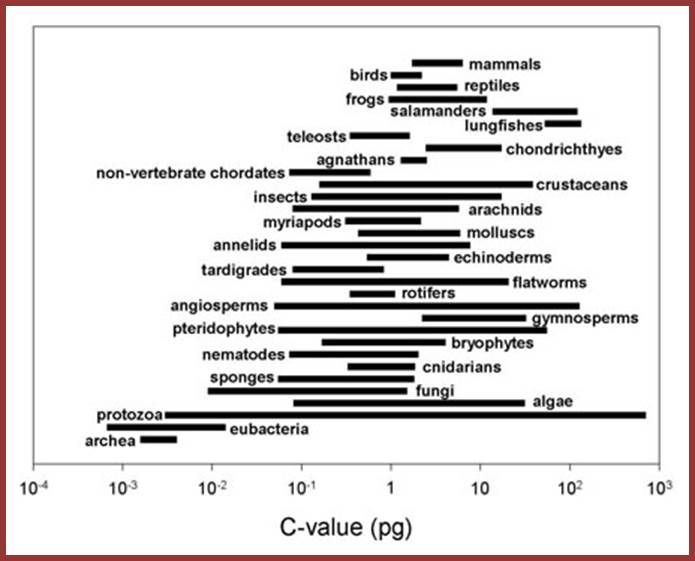
Note: the groups in this figure are arranged along made-up "scala naturae" to emphasize the lack of relationship between genome size and intuitive notions of organismal complexity -- please do not construe this figure as an endorsement of a progressionist view of evolution!). Laurence A.Moran; http://sandwalk.blogspot.in/
Considering the above criteria, variation in the size of the genome within a group or between closely related groups, it is not possible to explain why organism have so much of excess DNA than that can be accounted for. Is there any large-scale duplication of genes into multiple copies? Is there any DNA just that exists without any function? Is there any possibility that during evolution many genes have been duplicated and some have lost their functional abilities and exist as molecular fossils? Is there any possibility of promiscuous enmass transfer of the genome from other members? There are innumerable cases, especially plants, where by duplication of haploid chromosomes into diploids, triploids and polyploids, either by auto polyploidy or heteropolyploidy, the genome size has increased many fold, which has also created some variation in the size and also created new varieties or in extreme cases new species. Duplication of genomes in developing ploidy is different from the excess of DNA that is found in a haploid genome with no specific function. The question to be answered is from where this DNA has come, what DNA is it, how it has integrated and many such questions have to be answered.
It is important to know and solve this puzzle, by assessing how much DNA is actually used for coding and how much of this DNA is redundant, duplicated, and how many copies of each of them exist in a given genome. How much of DNA has no function and what is the size of these components and how many copies of each of these segments are made up off. If the DNA that doesn’t have any function or cannot be accounted for then what is it doing?, where is it located within the genome? Now techniques are available to determine the quality and quantity of protein coding genes, ribosomal RNA, tRNA and other small molecular weight nc RNA genes. It is possible to quantitate total DNA that codes for the above said components and also determine that DNA which doesn’t code for any functions. It is also possible to find out the number of each of the said category of DNA complexes.
It is also possible to estimate the number of genes expressed in tissue specific manner and to find the number of genes expressed as housekeeping genes. Even those genes that are expressed as housekeeping or tissue specific manner or expressed in response to age, stage of growth and development or response to stimuli, can be determined qualitatively and quantitatively.
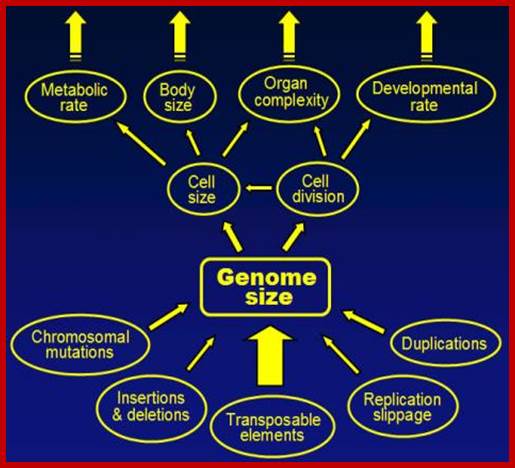
During the course of evolution, the existing genome is added with different components by various means, thus the genome size has increased, sometime abnormally. This increase in the genome size is used for various features like increased metabolism, cell and body size, organ complexity, developmental complexity and intelligence and survival ability.
Solving the Paradox:
The techniques used are DNA: DNA-DNA hybridization, DNA: RNA hybridization and DNA, RNA and Protein micro array. Hybridization techniques can be used to understand reassociation kinetics, which provides methods for quantification of each kind of DNA or RNA. The methodology used is simple, where the DNA is fragmented to the required size and fragments of the genome are taken and heated to its melting temperature ie.90°C (Tm) or more in a defined solution at a particular pH. When DNA fragments melt and strands separate completely, then they are allowed to renature or hybridize or anneal to each other in sequence specific manner at temperature 25° C less than its melting temperature. The temperature at which dsDNA strands melt is called Tm (melting temperature), which varies from DNA from one species to the other and again it depends upon the content GC or AT. DNA with more of GC content melts at higher temperature than the DNA with more of ATs.
Free nucleotides when used for measuring its quantity by means spectrophotometer, at UV wavelength 260nm, because of the presence of nitrogen bases with heterocyclic rings and double bonds, the nucleotides absorb light; the property is termed as chromacity. If the same amount of nucleotides in the form of polynucleotide chains such as dsDNA or DNA-RNA hybrids shows 40% less absorption, it is called Hypochromacity. On the contrary, if the ds stranded DNA is melted into single stranded DNA, its OD increase by 40%, which is called Hyperchromacity. If one OD of dsDNA at 260nm, gives a quantity of 50ug/ml, but one OD of ssRNA gives 40ug/ml, this is because in ds DNA not all the nucleotides are exposed or oriented for absorption of light and some are hidden from the light, because the DNA is in coiled state, while RNA being single stranded structure, all of its nucleotides are exposed to light, so it shows higher OD or it is hyperchromatic. This phenomenon can be exploited in quantification of ss DNA and ds DNA in dissociation and reassociation experiment; the same can also be used to study kinetics of reassociation, which provides valuable data.
Melting Point of DNA:
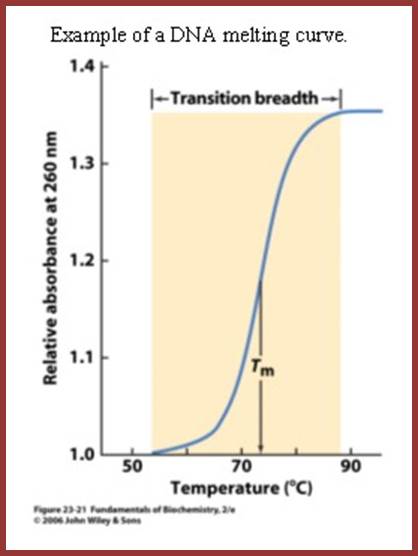
http://classes.biology.ucsd.edu/
Curve shows the Tm at which melting starts and ends
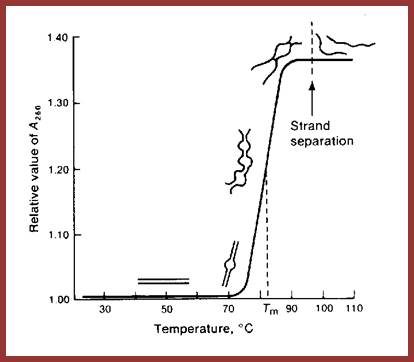
http://bioscience.egloos.com/
This Tm curve shows, above and below, how and at what temperature the ds DNA melts into single stranded DNA
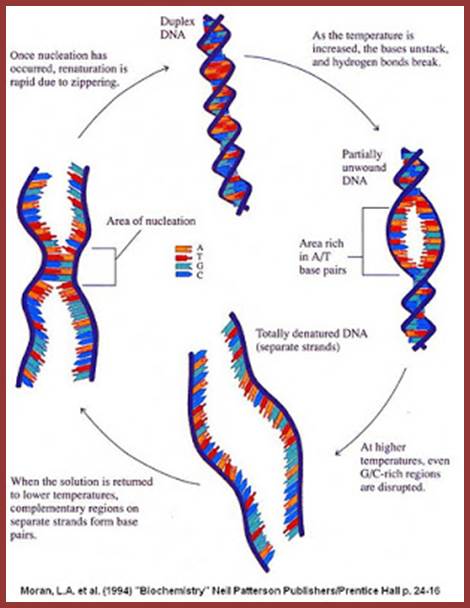
http://sandwalk.blogspot.in/

Thermal denaturation (DNA melting); http://www.entelechon.com/
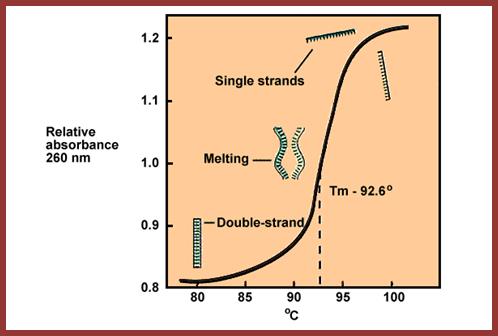
Tm curve; https://www.dkfz.de
When a given DNA, in an ionic solution with a specific pH, heated, it slowly melts into ssDNA, then the melting is fast as the temperature is raised. The temperature at which it completely melts, it is called melting point of DNA. But the term Tm is used to define the temperature at which half of the DNA is melted. The Tm of the DNA varies depending upon the G+C content, if the G+C content is more than A+T, the Tm is more and the reverse of it is true to A=T rich DNA. DNA-DNA hybridization, DNA-RNA hybridization combined with sequencing it is possible to relate one species to the other- i.e phylogeny.