Protein Degradation:
Protein stability and Protein turnover:
- Cells continuously synthesize a variety of proteins in cell specific manner and stage specific manner, all the time. In the same way, proteins, which are not required or those, which are abnormally folded or partially degraded or not properly made, or damaged in the process or aged, are removed by degradation. This process of synthesis and degradation of each kind or kinds is called protein turnover. Scientists call this process as “From Cradle to Grave”.
- The rate of turn over varies from one protein species to the other species. Bacterial proteins half life ranges from as short as 20 seconds to 2-4 minutes. Some have half-life of few minutes and some have 100 days or more. Proteins turnover refurbish the cells with fresh and required new proteins and old and not required proteins are removed (salvage pathway), otherwise they cause great injury to cells.
- To illustrate the point, at a particular stage of development certain specific proteins are required only for a particular duration of time. Once their function is over, they are not required any more. Continued presence of them is disastrous to that cell, so they have to be degraded in stage specific manner. Some such proteins are synthesized only once in the lifetime of a specific cell or an organism.
- More than this, proteins permits metabolism of a cell to be in active state and can regulate their cellular processes. Among all the cells RBCs, lymphocytes, neuronal cells, muscle cells, liver cells have long life and among cell organelles ribosomes and nuclei have long life span.
- Some cell type produce their proteins, which are specialized for the said cells, they have specific shape and functions. Such proteins have to be maintained in the required amounts and the aged or damaged cell specific proteins have to be replenished in required amounts all the time.
- Depending upon the extra cellular stimulus, certain genes are expressed transitorily and their products perform their function and then they degraded. A variety of intracellular stimuli or inter cellular events in one part of the body or an organ can provide a stimulus for which certain set of proteins are produced only for short period of time and they have to be degraded in required time.
- There are several thousands of proteins, which are continuously synthesized and degraded throughout the life of the cell, they are housekeeping proteins, though their turnover is slow, but operate all the time.
- Occasionally, due to distress, some cells commit suicide called apoptosis. In certain situations the self-inflicted death may ensue. Developmentally certain cells undergo progressive degeneration and death. All these events require, on one hand synthesis of proteins and on the other hand degradation of proteins; some specific and some general.
- Proteins that are not proper or contain undesirable adducts are subjected immediate degradation for they will be marked for destruction. Example if the hemoglobin with a Valine analog alpha amino B-chlorobutyrate is incorporated and expressed by recombinant methods, the said protein lives for a very short period of time. The cell recognizes the protein as undesirable and not good for it, so it is marked and destroyed. This form of hemoglobin has half-life of 10 minutes or less, where as the normal hemoglobin has a life span of more than 120 days or as long as the RBCs live; in fact cartilage proteins and crystalline (eye lens) are long lived proteins.
- Even in Bacterial cells, proteins that are not required are immediately degraded faster than eukaryotic cells. In fact the turnover of mRNA in prokaryotic cells is very fast; their half-life is just 2 to 3 minutes. For example, the mutant of B-Galactosidase is degraded within few minutes, while the normal b-Galactosidase remains undegraded for a long, long time.
- Internal milieu of cells is highly reactive one. So besides mutations, errors during protein synthesis, in such reactive environments, proteins are subjected to modifications, and become abnormal. The presence of such proteins is not compatible for cellular functions, so they have to be removed, and the only method to do is either by proteolysis or by excretion of them or by both.
- There is no in-built repair process or mechanism for proteins, but such mechanisms exist for DNA repair, which it should be for it is the master molecule that codes for proteins.
- If error prone proteins are produced, they have to be removed and new ones can be made, but it is not case with that of DNA, which is the master molecule of life, if an error occurs, it cannot be removed and new DNA cannot be replaced, only it can be repaired if possible.
- Different proteins are degraded at different rates, some are degraded fast and some are degraded slowly.
From the cradle to the grave: molecular chaperones that may
choose between folding and degradation:
Jörg Höhfeld, Douglas M. Cyr &
Cam Patterson.

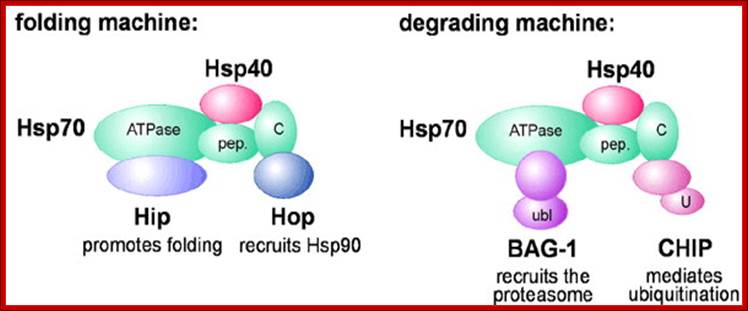
Hsp70 and Hsp90 in protein folding and degradation: An initial decision to fold or degrade a Hsp70- or Hsp90-associated substrate protein may be reached through competition between a 'productive' cofactor such as Hop, and the ubiquitin ligase CHIP. During the folding process, Hsp70 and Hsp90 may co-operate with other chaperone proteins (termed partner chaperones); HOP-Hsp70-Hsp90 Organizing Protein; CHIP-Ubiquitin ligase constitutive heat shock cognate 70 interactive protein.; From the cradle to the grave: molecular chaperones that may choose between folding and degradation; http://embor.embopress.org/
On the degradation pathway, Carboxy terminus of Heat shock Interacting Protein (CHIP) associates with Hsp70 or Hsp90 via its Tetratricopeptide TPR repeats, and at the same time recruits E2 ubiquitin conjugating enzymes of the Ubc4/5 family to the chaperone complex. This may involve binding of the E2 to the U-box of the cofactor (U). In conjunction with E2, CHIP mediates ubiquitin attachment to the chaperone substrate and induces its targeting to the Proteasome for degradation. The targeting process may be facilitated by an ubiquitin domain protein (UDP), such as BAG-1, which binds to Hsp70 and utilizes its ubiquitin-like domain (ubl) for proteasomal association C terminal Hsc70 interacting protein-Chip. In protein degradation besides lysosomes, another method of protein degradation was found in2004 by Aaron Ciechanover, Avram Hershco and Irwin Rose; it is now popularly called Proteasome a giant killer of proteins. Proteasomes of 26S size are found in archaea and eukaryotes; in eukaryotes they are located in cytoplasm and even in the nucleus. Some are also found in some bacteria, ex. Anbu 20s Proteasome.
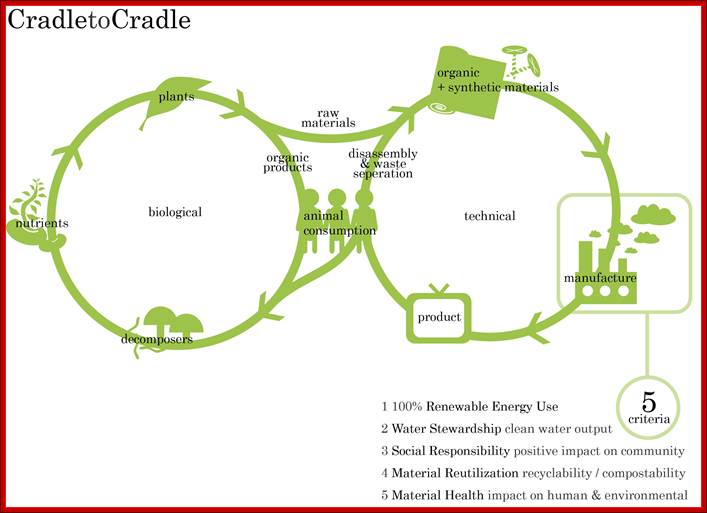
Initially defined by McDonough and Braungart, the Cradle to Cradle or Is it Cradle to Grave? http://en.wikipedia.org/;
Overview of Golgi and protein processing:

The diagram at the left summarizes the flow of proteins between the compartments of the endomembrne system. In the initial section of this unit we will consider the transfer of proteins from ER to golgi (red arrow) and the reverse (retrograde) transfer of receptors and proteins destined for retention in the ER compartment (blue arrow). The non-highlighted portion of the figure shows the flow of proteins in the secretory, lysosomal and endocytotic pathways that will be considered later. Click on image to enlarge. http://www.zoology.ubc.ca/
Half life of some mRNAs
|
mRNA species |
Half-life (Hours) |
|
|
β-actin (nonmuscle) |
60 |
|
|
Cytochrome P450 |
36 |
|
|
Fatty acid synthetase |
48-96 |
|
|
Glu-6-Phosphate dehydrogenase |
15 |
|
|
Glyeraldehyde 3-P dehydrogenase |
75-130 |
|
|
Heat Shock Proteins (70hps) |
2.0 |
|
|
Insulin receptor |
9 |
|
|
Thymidine kinase |
2.6 |
|
|
Tubulin |
4 -12 |
|
|
Tyrosine aminotransferase |
2.0 |
|
|
Glucocorticoid receptor |
2.3 |
|
|
Ornithine decarboxylase |
0.5 |
|
|
Most bacterial mRNA's |
< 3.0 minutes |
Half Life of Some proteins:
|
Protein |
Half-life in hours |
|
Short lived: |
|
|
Ornithine carboxylase |
0.2 hrs |
|
RNAP-1 |
1.3 |
|
Serine dehydratase |
4 |
|
PEP carboxylase |
5 |
|
Long lived: |
|
|
Aldolase |
118 |
|
GAPDA |
130 |
|
Cyt.B oxidase |
130 |
|
Lactate dehydrogenase |
130 |
|
Cyt.C oxidase |
150 |
|
Crystalline |
70-80 yrs |
Half Life of Some Proteins: N-terminal Sequence Dependent:
|
N-terminal sequence |
Half life |
|
N-M.S.A.T.V.G |
20 hrs |
|
N-I.E- |
30 min |
|
N-Y.Q- |
10 min |
|
N-L.D.K or R- |
2 min |
|
PEST, N degradons |
5-8 hrs |
|
|
|
|
|
|
N-terminal sequences involved in degradation are called N-degradons.
Stretches of PEST sequences which are rich in proline (P), glutamate (E), serine (S), and threonine (T) (along with a lesser extent, aspartic acid) serve as a destruction signal (so called "PEST sequences").
Ubiquitination of mitotic cyclins is mediated by a small NH2-terminal motif known as the "destruction box" or “D-box” (Glotzer et al., 1991). The minimal motif is nine residues long with, the following consensus sequence: R-A/T-A-L-G-X-I/V-G/T-N. The destruction box, while either phosphorylated or ubiquitylated also serve as a binding site for the ligase subunit of the APC/cyclosome complex.
N-glycans were recently found to act as ubiquitination signaling molecules. It was recently demonstrated that Fbx2, component of large SCF-type E3 ubiquitin ligase complex specifically binds N-linked glycoproteins and ubiquitinates them, leading to degradation via the endoplasmic reticulum associated protein degradation (ERAD) pathway.
The molecular chaperones are known to bind misfolded or unfolded proteins to prevent protein aggregation. They either catalyze the refolding of the protein through an ATP-dependent mechanism (if feasible) or target these misfolded proteins for ubiquitination. CHIP (C-terminus of Hsc70-interacting protein) is an excellent example of U-box E3 ligase family as it targets the misfolded proteins.
Additionally, studies have revealed that a specific ubiquitin ligase recognizes phosphorylated IKBα (pIKBα) through a short peptide stretch, composed of 6 aa motif ( e.g., DS(PO3)GXXS(PO3)).
Prot-Pharam tool: The Proteomics Protocols Handbook, Humana Press (2005). Half-life of proteins with a.a at N end of the proteins.
|
Amino acid |
Mammalian |
Yeast |
E.coli |
|
Ala |
4.4hr |
>20hr |
>10hr |
|
Arg |
1hr |
2min |
2min |
|
Asn |
1.4hr |
3min |
>10hr |
|
Asp |
1.1hr |
3min |
>10hr |
|
Cys |
1.2hr |
>20 |
10hr |
|
Gln |
0.8hr |
10min |
>10hr |
|
Glu |
1hr |
30mn |
>10hr |
|
Val |
100hrs |
20hrs |
10hrs |
|
Gly( G) |
30hr |
20 |
10hr |
|
His |
3.5hr |
10min |
10hr |
|
Ile |
20hrs |
30min |
10hrs |
|
Leu |
5.5hrs |
3min |
2min |
|
Lys |
1.3hr |
3min |
2min |
|
Met |
30hr |
20 |
10 |
|
Phe |
1.1hr |
3min |
2min |
|
Pro |
20hrs |
2phrs |
|
|
Ser |
1.9hrs |
20 |
10hrs |
|
Thr |
7.2hrs |
20hrs |
10hrs |
|
Trp |
2.8hrs |
3min |
2min |
|
Tyr |
2.8hrs |
10min |
2min |
- Enzymes or proteins in general with N-terminal sequence such as N-
M.S.A.T.V.G. or C are found to be long lived. This conclusion has been arrived by studying the N-terminal sequences of about 209 or more enzymes. The N-end rule pathway is now emerging as a major cellular proteolytic system, in which the majority of proteins are born with or acquire specific N-terminal degradation determinants through protein-specific or global post-translational modifications; Nature review 2011
- Proteins containing N-terminal a.a Phe, Leu, Asp, Lys, or Arg have half lives of 3 min or less.
- Those proteins, which have Proline, Glutamic acid, Ser or Threonine residues called PEST sequences are degraded rapidly?
· Majority of the regulatory proteins, in the sense, which control or regulate a transitory event or processes, for example response to a stimulus, are degraded very quickly; such rapid turnover greatly facilitates the cell to respond to changes in nutrition, temperatures, pH, hormones, etc. Interestingly some of the cytoplasmic proteins are degraded quickly, because their half-life is dependent on their N-terminal amino acid sequences.
Degradation Pathways:
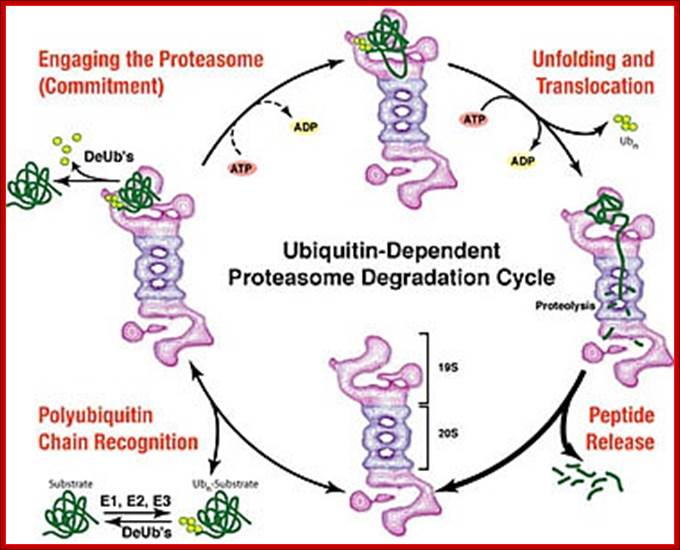
Most of the cells employ two pathways; one is lysosomal mediated and the other is ATP dependent Proteasome mediated method.

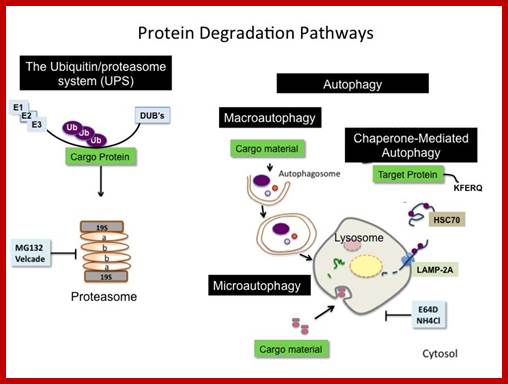
Top fig; There are two major fundamentally different mechanisms by which cells degrade proteins for turnover and recycling purposes: the lysosome and the proteasome. Our research uses a creative combination of pharmacological, biochemical and genetic approaches to rigorously investigate the biological significance of these degradation mechanisms in normal and cancer cells. http://ki.se/
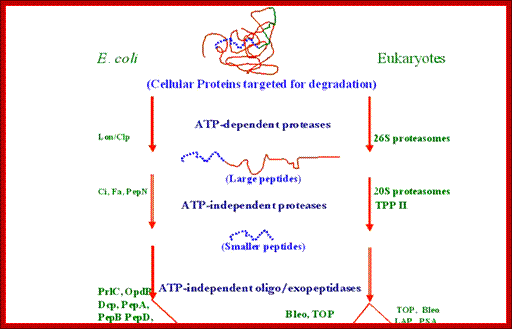
Bottom fig; The figure depicts the molecules involved in the intracellular protein degradation pathway. Cytosolic protein degradation are categorized into four step : 1) protein targeted for degradation are initially unfolded into polypeptides by ATP dependent protease belonging the Lon/Clp family in bacteria or 26S proteasome in eukaryotes. 2) After unfolding these enzymes also make an initial cut in the polypeptide using their endoproteolytic property. 3) The longer peptides released by the above proximal enzymes are further trimmed into smaller peptides by the action of endopeptidase , tripeptidyl � and dipeptidyl peptidases. 4) Finally, these oligopeptides are acted on by amino or carboxy peptidases to release free amino acids. http://biochem.iisc.ernet.in/
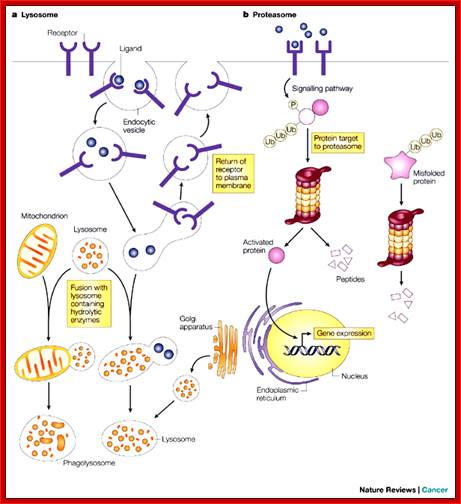
The two main routes of intracellular protein degradation are the lysosomes and the proteasomes. a | Lysosomes are cytoplasmic, membrane-bound vesicles that enclose proteases and hydrolytic enzymes. They degrade extracellular and transmembrane proteins that are taken up by endocytosis — the process by which the cellular plasma membrane invaginates and breaks off internally to transport materials into the cell — and participate in autophagocytosis by fusing with phagosomes. b | Proteasomes are primarily involved in degrading intracellular proteins. These might be targeted by phosphorylation following activation of signalling pathways, or recognized because they are misfolded. The proteins are targeted for degradation by their ubiquitin tag. Julian Adams;http://www.nature.com/
Lysosomes and Lysosome mediated method;
Lysosomes are bags of digestive enzymes of 50 or more kinds and most of them degrade a variety of compounds in nonselective manner. The proteases found are a family of enzymes called Cathepsins, which are proteases of specific class. Lysosome by using its ion transport system, maintain intra-vacuolar liquid at PH 5.0 and this pH renders the lysosomal enzymes inactive. Chloroquine is an antimalarial drug, and it inhibits lysosomal activities. Chloroquine consists of two hexagonal rings with N in one of the rings linked to, two rings-NH-CH (-CH3)-(CH2) 3-N- (C2H5) 2. It is a weak base, freely penetrates across the membrane and accumulates in charged form and inhibits lysosomal enzymes. Cathepsins inhibitors like Antipain (an antibiotic), inhibits protein degradation. Lysosome, in general, recycle intracellular constituents by fusing with autophagic vacuoles and break down its contents. Many of the ingested particulates as phagocytotic or endocytotic vesicles are fed to lysosomes via late endocytotic vesicles. These organelles are not just meant for protein degradation, but they are also meant for degradation of carbohydrates, lipids and other macromolecules, which can be used as reserve energy or food source.
Some important enzymes found within lysosomes include:
- Lipase, which digests lipids
- Amylase, which digests amylose, starch, and maltodextrins
- Proteases, which digest proteins
- Nucleases, which digest nucleic acids
- Phosphoric acid monoesters
Lysosomes are frequently nicknamed "suicide-bags" or "suicide-sacs" by cell biologists due to their role in autolysis. Lysosomes were discovered by the Belgian cytologist Christian de Duve in the 1950s.
Functions:
“Lysosomes are the cell's waste disposal system “Garbage Cleaner” and can break up anything. They digest almost everything. They are used for the digestion of macromolecules from phagocytosis (ingestion of other dying cells or larger extracellular material, like foreign invading microbes), endocytosis (where receptor proteins are recycled from the cell surface), and autophagy (where in old or unneeded organelles or proteins, or microbes that have invaded the cytoplasm are delivered to the lysosome). Autophagy may also lead to autophagic cell death, a form of programmed self-destruction, or autolysis, of the cell, which means that the cell is digesting itself.
Other functions include digesting foreign bacteria (or other forms of waste) that invade a cell and helping repair damage to the plasma membrane by serving as a membrane patch, sealing the wound. In the past, lysosomes were thought to kill cells that were no longer wanted, such as those in the tails of tadpoles or in the web from the fingers of a 3- to 6-month-old fetus. While lysosomes digest some materials in this process, it is actually accomplished through programmed cell death, called apoptosis. Different storage diseases may have similar clinical features, thus it may be necessary to measure a number of different enzyme activities prior to finding the one deficient in a particular patient. The Metabolic Laboratory of the Greenwood Genetic Center offers a lysosomal enzyme panel that includes the enzymes listed below. These tests can also be ordered on an individual basis, if desired”.
α-fucosidase
(Fucosidosis)
α-Galactosidase
(Fabry disease)
α-iduronidase
(Hurler syndrome; MPS I)
α-mannosidase
(α-mannosidosis)
α-neuraminidase
(sialidosis) *available on fibroblasts only
β-galactosidase
(GM1 gangliosidosis)
β-glucosidase
(Gaucher disease)
β-glucuronidase
(Sly syndrome; MPS VII)
β-mannosidase
(β-mannosidosis)
To accomplish the tasks associated with digestion, the lysosomes use some 40 different types of lysosomal enzymes, such as glycosidase, protease, acid phosphatase, sulfatases and lipases. Lysosomal enzymes are synthesized in the endoplasmic reticulum and modified in the Golgi apparatus. Lysosomal enzymes are tagged for lysosomes by the addition of mannose-6-phosphate label. A deficiency of any one of these lysosomal enzymes will lead to lysosomal storage diseases, such as Tay-Sachs disease and Pompe's disease. Lysosomal enzymes are also known to be involved in cancer processes.
Degradation of short lived or abnormal proteins has nothing to do with lysosomal hydrolytic activities. Prolonged starvation, leads to excessive lysosomal activity leading to digestion of essential enzymes and regulatory proteins, this effect is devastating to cells and tissues. Though lysosomes have nonselective degradation, yet in certain situations lysosomal enzymes exhibit selective proteolysis of those, which contain a pentapeptide sequence of K.F.E.R.Q (lys.phe.glu.arg.gln) or related sequence. In fasting animals, such proteins are lost from tissues like liver and kidneys, but not from brain and testis tissues. Certain cytosolic factors bind to such proteins that have those sequences and deliver them to lysosomes for digestion. The factors that recognize and bind to such proteins are 73 KD PrP 73. The PrP 73 is member of heat shock proteins.

The acid hydrolases are hydrolytic enzymes that are active
under acidic conditions. The lumen is maintained at an acidic pH by an H+ ATPase in the membrane that pumps H+into
the lysosome. ;http://homepages.uc.edu
http://www.sivabio.50webs.com
The acid hydrolases are hydrolytic enzymes that are active under acidic conditions. The lumen is maintained at an acidic pH by an H + ATPase in the membrane that pumps H + into the lysosome; http://www.sivabio.50webs.com/
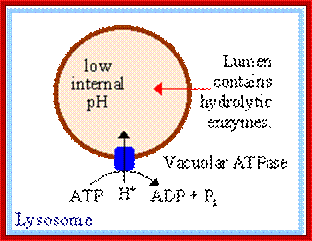
ATPase pumps maintain acidic state in lysosomal fluid; https://www.rpi.edu

http://slideplayer.com; Plant cells have central vacuole which acts not only as lysosome but also osmoregulator.
Plant central vacuole is store house many enzymes which are used; https://www.haikudeck.com
Lysosome Mediated Effects:
|
Disease
|
Causes, why/how |
|
Rheumatoid arthritis: |
It is due to the release of lysosomal enzyme into extra cellular region causing breakdown of a number proteins and a number of cell types, it that causes inflammation and pain.
|
|
Diabetes mellitus |
Stimulate lysosomal breakdown of a variety of proteins.
|
|
Muscle wastage; |
Due to disuse, again due to excessive lysosomal activity. |
|
Traumatic injuries |
Again this is due to increased lysosomal activity. |
|
Regression of uterus after child birth |
Many muscular organs are reduced from few kg to few grams.
|
|
Regression of tadpole tail
|
During morphogenesis, it is the activity of lysosomes by digestion, regress the tadpole tail from few inches to nothing in adult frog.
|
Lysosome Biogenesis:
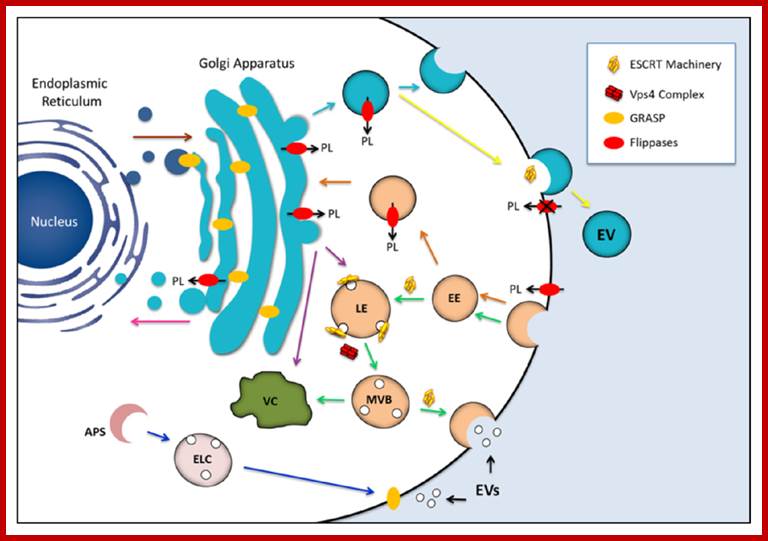
Golgi complex from Trans-membranes buds off vesicles containing lysosomal enzymes; they are sorted. Potential participation of components of the endosomal sorting complex required for transport (ESCRT) machinery, GRASP and flippases in the biogenesis of fungal extracellular vesicles (EVs). The similarities between EVs produced by fungi and mammalian exosomes suggest that ESCRT machinery is required for formation of the fungal compartments (green arrows). Maturation of the late endosome (LE) is accompanied by membrane invagination, giving origin to small intraluminal vesicles and multivesicular bodies (MVB). The ESCRT machinery is recycled through the activity of the Vps4 protein complex. MVB may be directed to vacuolar (VC) degradation pathways, but also to fusion with the plasma membrane, releasing exosomes to the extracellular milieu now receiving the name exosomes. GRASP, a regulator of unconventional secretion by mechanisms that are putatively linked to EV release, was first identified as a structural component of the Golgi cisternae. Alternative roles included tethering activity for endosomal or lysosomal compartments and/or regulation of autophagy-related mechanisms (blue arrows). GRASP may also localize to the plasma membrane, mediating the release of exosomes to the extracellular space. Finally, GRASP can also participate in docking or fusion events involving vesicles originating at the Golgi, thus facilitating anterograde transport through the early secretory pathway (brown arrow). Flippases are involved in vesicle biogenesis through phospholipid translocation across the lipid bilayers. These enzymes can regulate endocytosis at the plasma membrane level (orange arrows) and also drive the formation of exocytic vesicles (light blue arrows). Flippases can also participate in protein trafficking between the trans-Golgi network and endosomal compartment or between the trans-Golgi network and vacuoles (purple arrows). It has been also proposed that flippases may regulate the retrograde transport pathway from the Golgi apparatus to the ER (pink arrow), as well as vesicle budding at the plasma membrane level (yellow arrow). The possibility that cellular pathways regulated by endosomal proteins, GRASPs and flippases are interconnected cannot be ruled out, as previously described for other unconventional secretory pathways [165]. Most of the mechanisms proposed here have been implicated with the physiology of yeast cells, although they also participate in pathways required for molecular degradation and / or export in other eukaryotes. (PL) phospholipid; (EE) early endosome; (LE) late endosome; (MVB) multivesicular bodies; (APS) autophagosome; (ELC) endosomal/lysosomal compartment; (VC) vacuole. http://www.mdpi.com
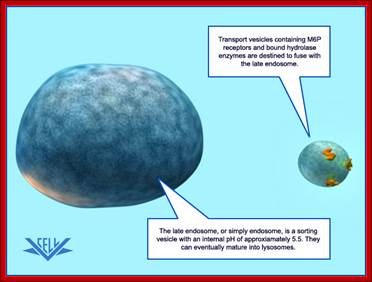
Purified Lysosome; www.vcell.science
The transport vesicle marked and loaded with lysosomal enzymes from the trans-Golgi moves toward early endosomes and fuse with the same; the early endosomes derived from the PM fuse with late endosome, which can eventually mature into a lysosome.

Three pathways to degradation in lysosomes; cytochemistry.net/cell-biology/lysosomes.htm
Each pathway leads to the intracellular digestion of materials derived from a different source. The compartments resulting from the three pathways can sometimes be distinguished morphologically - hence the terms "autophagolysosome," "phago-lysosome," and so on. Such lysosomes, however, may differ only because of the different materials they are digesting.

cytochemistry.net/cell-biology/lysosomes.htm
The transport of lysosomal hydrolases to lysosomes:
The precursors of lysosomal hydrolases are covalently modified by the addition of mannose 6-phosphate groups (M6P) in the cis Golgi network. They then become segregated from all other types of proteins in the trans Golgi network because a specific class of transport vesicles (called clathrin-coated vesicles) budding from the trans Golgi network concentrates mannose 6-phosphate-specific receptors, which bind the modified lysosomal hydrolases. These vesicles subsequently fuse with late endosomes. At low pH in the late endosomes hydrolases dissociate from the receptors, which are recycled to the Golgi apparatus for further rounds of transport. In late endosomes the phosphate is removed from the mannose on the hydrolases, further ensuring that the hydrolases do not return to the Golgi apparatus with the receptor
Lysosomes in general are meeting places in which several streams of intracellular traffic converge. Digestive enzymes are delivered to them by a route that leads outward from the ER via the Golgi apparatus, while substances to be digested are fed in by at least three paths, according to their source.
Of the three paths to degradation in lysosomes, the best studied is that followed by macromolecules taken up from the external medium by endocytosis. In brief (for the details will be discussed later), the endocytosed molecules are initially delivered into small, irregularly shaped intracellular vesicles called early endosomes. From these, some of the ingested molecules are selectively retrieved and recycled to the plasma membrane, while others pass on into late endosomes. Here, by fusion of two streams of transport vesicles, the materials coming in for digestion first meet the lysosomal hydrolases coming out from the Golgi apparatus. The interior of the late endosomes is mildly acidic (pH~6), and it is thought to be the site where the hydrolytic digestion of the endocytosed molecules begins. Mature lysosomes form from the late endosomes, although it is not known precisely how this occurs. During the conversion process some distinct endosomal membrane proteins are lost, and there is a further decrease in internal pH.
A second pathway to degradation in lysosomes is used in all cell types for disposal of obsolete parts of the cell itself - a process called autophagy. In a liver cell, for example, an average mitochondrion has a lifetime of about 10 days, and electron microscopic images of normal cells reveal mitochondria engulfed lysosomes (presumably digesting) as well as other organelles. The process seems to begin with the enclosure of an organelle by membranes derived from the ER, creating an autophagosome, which then fuses with a lysosome (or a late endosome). The process is highly regulated and selected cell components can somehow be marked for destruction during cell remodeling: the smooth ER that proliferates in a liver cell in response to the drug Phenobarbital is selectively removed by autophagy when the drug is withdrawn.
As we discuss later, the third pathway that provides materials to lysosomes for degradation occurs mainly in cells specialized for the phagocytosis of large particles and microorganisms. Such professional phagocytes (macrophages and neutrophils in vertebrates) engulf objects to form a phagosome, which is then converted to a lysosome in the manner described for the autophagosome.
Plant central vacuole- a massive Lysosome,www.slideplayer.com/slide
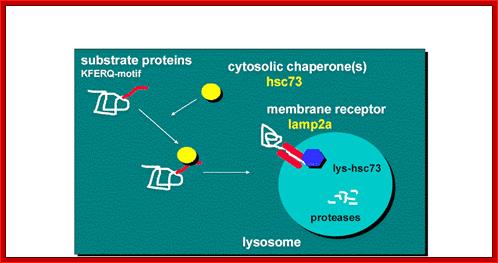
Lysosomal
proteins, marked are facilitated by chaperones, bind to lysosomal membrane
receptors and Lysosomal proteins are thus transported
https://www.einstein.yu.edu
“Chaperone-Mediated Autophagy (CMA) is responsible for the degradation of approximately 30% of cytosolic proteins in tissues such as liver, kidney and in many types of cultured cells.
This lysosomal mechanism of protein degradation is mainly activated in conditions of stress such as nutrient deprivation or exposure to different toxin compounds. Under those conditions, the substrate proteins for this pathway, all those containing a motif biochemically related to the pentapetide KFERQ , are selectively recognized by a cytosolic chaperone, the heat shock cognate protein of 70 kDa (hsc70). The interaction between the chaperone and the substrate in the cytosol targets the complex to the lysosomal membrane where it binds to the lysosome associated membrane protein type 2A (LAMP-2A) that acts as a receptor for this pathway. A second chaperone, the lysosomal hsc73 (lys-hsc70), is required for the complete translocation of the substrate protein into the lysosomal matrix where it is completely degraded by the lysosomal proteases”.
Proteasome method:
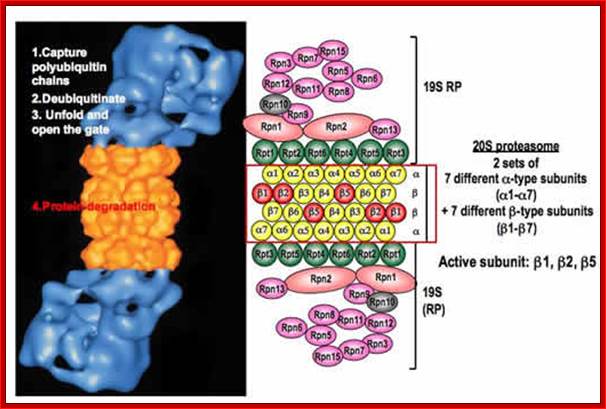
Proteasome is a huge complex of proteins that degrades most of the marked protein for degradation. Its size itself is 26S, almost the size of small ribosomal subunit. This protein complex is fed with marked proteins. The marking is done by two other processes called ubiquitination and sumoylation; (functions of them are different. ). Bacterial proteasome is called ‘Anbu’ found in proteobacteria.
Proteasome is made up of several protein subunits coded for by at least 14 genes (7alpha and 7beta). The size of proteasome is 100x160 °A, consists of 2-alpha rings and 2-beta rings (a-b-b-a); each ring consists of seven different subunits having molecular weight of 20 to 30kDa. The lid consists of seven subunits and central two rings also contain seven subunits each. Each barrels are capped by 19s proteins.
Eukaryotic Proteasome:

Structure of the proteasome. The proteasome is a modular structure. One or two regulatory particles (RP) attach to the outer surface of the core particle (CP). The CP is made up of four heptameric rings: two outer identical α-rings and two identical inner β-rings. Each ring is made of seven distinct homologous subunits. Certain β-subunits contain the protease active sites facing inward into the sequestered proteolytic chamber. Upon interferon-γ induction, three β-subunits can be replaced by βi (LMP) homologs that alter the proteolytic specificities of the proteasome. The 19S RP is comprised of two eight-subunit subcomplexes, the lid, and the base. The base that contains all six proteasomal ATPases attaches to the α-ring of the CP. The lid can disassociate from the proteasome, resulting in a truncated base-CP complex. Rpn10 can interact with either the lid or the base and stabilizes the interaction between the two. Rpn10 is also found outside of the proteasome. Numerous associated proteins and auxiliary factors, such as chaperones and components of the ubiquitination machinery, can interact with the RP. Alternative regulatory complexes can also attach to the surface of the α-ring; Michael H. Glickman , Aaron Ciechanover; http://physrev.physiology.org/
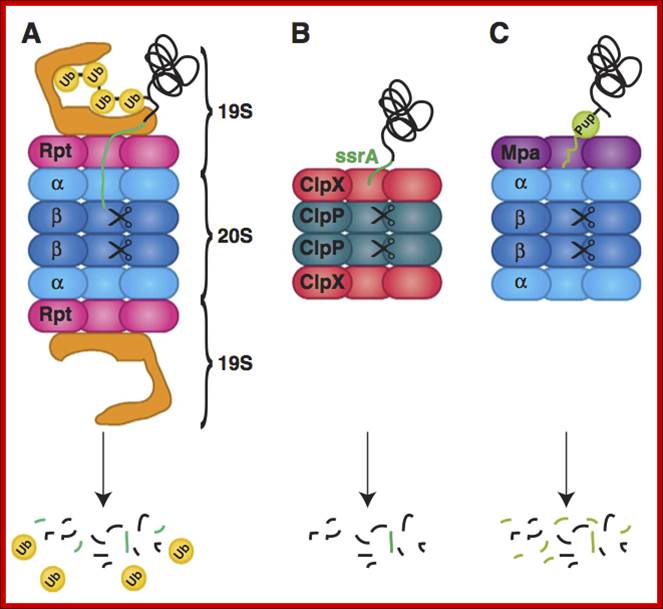
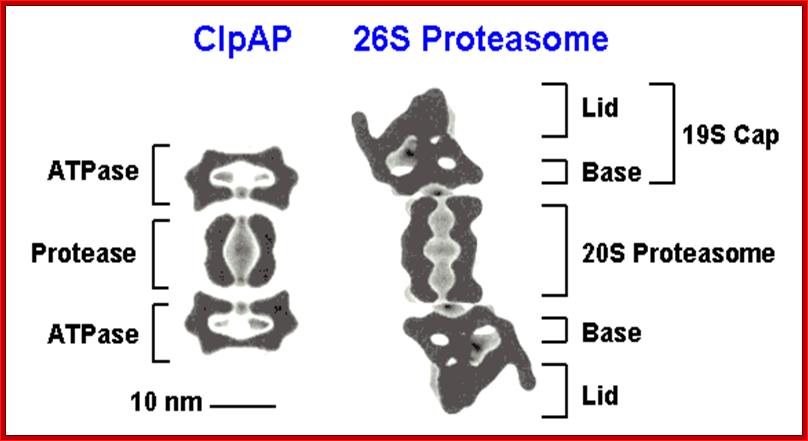
Figure; Comparison of different classes of ATP-dependent proteases, shown as a side-on cross-section. (A) The eukaryotic 26S proteasome, composed of a 20S core particle (blue a and b subunits) flanked by 19S regulatory subunits (magenta and orange). The 19S subunits bind to the substrate through the covalent ubiquitin modification (yellow) and unfold it by pulling on the unstructured initiation site (bright green). Ubiquitin is removed to be recycled during the degradation process. (B) The bacterial protease ClpXP, composed of rings of the protease ClpP (dark green) and the ATPase motor ClpX (red). ClpX binds to the degradation signal, in this case the ssrA peptide sequence (green), which also serves as the site for the initiation of degradation. (C) The actinobacterial proteasome, consisting of a 20S core particle similar to that of the 26S proteasome, and a single ring of the ATPase Mpa (purple). Mpa binds to the substrate through the covalent Pup modification (light green). Pup has an N-terminal unstructured region, which serves as the site for the initiation of degradation, leading to complete degradation of Pup. (D) Archaeal proteasome is most closely related to the eukaryotic proteasome, with core alpha and beta subunits and Rpt-like AAA ATPase subunits (termed PAN for Proteasome Activating Nucleotides) involved in protein unfolding; Cecile M. Pickart & Robert E. Cohen

Cartoon representation of a proteasome: Its active sites are sheltered inside the tube (blue). The caps (red; in this case, 11S regulatory particles) on the ends regulate entry into the destruction chamber, where the protein is degraded,. https://www.rpi.edu
Side view and Top view of the same schematic, illustrating the seven-fold symmetry of the rings. The mammalian complex is 26S with mol.wt of 2000kDa. This complex consists of 20S protein subunit and two 19s regulatory cap subunits. The size is~115Ax53A and entrance is 13A, Willow W. http://bioweb.uwlax.edu/
A schematic diagram of the proteasome 20S core particle (above) viewed from one side. The α subunits that make up the outer two rings are shown in green, and the β subunits that make up the inner two rings are shown in blue. Unlike active proteases, the proteasome is capable of degrading any protein, yet it has some specificity.
The key trick is autocompartimentación. The active sites of proteolytic 20S core particles (CPs for its acronym in English) are abducted from their cellular environment inside of this barrel-shaped sub complex. Proteins destined for degradation are marked by a polyubiquitin chain, a signal degradation that recognize regulatory particle 19S (RPS) that bind to one of the two ends or both of the CP [nuclear particle] to form the holocomplejo 26S.RPS [controlling particles] (i) recognize poliubiquitilados substrates, (ii) and recycled trimmed polyubiquitin chains, (iii) display substrates to be degraded, and (iv) opening the gate to the CP and aid in the transferring substrates within the CP. These tasks are performed by a complex machinery involving at least 19 different subunits, 6 AAA-ATPases (Rpt1-6 y) 13 non-ATPases (Rpn1-3-13 and Rpn15/Sem1p Rpn5
In structure, the proteasome is a cylindrical complex containing a "core" of four stacked rings forming a central pore. Each ring is composed of seven individual proteins. The inner two rings are made of seven β subunits that contain three to seven protease active sites. These sites are located on the interior surface of the rings, so that the target protein must enter the central pore before it is degraded. The outer two rings each contain seven α subunits whose function is to maintain a "gate" through which proteins enter the barrel. These alpha-subunits are controlled by binding to "cap" structures or regulatory particles that recognize polyubiquitin tags attached to protein substrates and initiate the degradation process. The overall system of ubiquitination and proteasomal degradation is known as the ubiquitin-proteasome system-Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko and Irwin Rose,2004.
Ubiquitination or Ubiquitylation:
An abnormal protein or proteins with errors, truncated or other non-performing wrongly folded or modified proteins, inside cellular milieu including ER, is a burden and can cause problems for the normal metabolic activities and health of the cells.
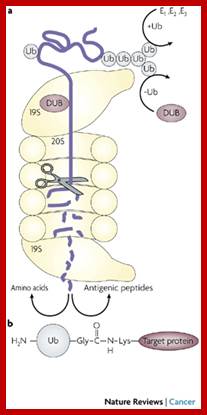
- Specialized proteins called ubiquitin mark such proteins; such marked proteins are fed into protein chewing monster machines called proteosomes.
Ubiquitin (originally, ubiquitous immunopoietic polypeptide) was first identified in 1975 as an 8.5-kDa protein of unknown function expressed in all eukaryotic cells. The basic functions of ubiquitin and the components of the ubiquitination pathway were elucidated in the early 1980s in a ground breaking work performed at Fox Chase Cancer Center by Aaron Ciechanover, Avram Hershko, and Irwin Rose ; Irwin Rose was awarded Nobel Prize 2004 in Chemistry for his work.
A diagram of ubiquitin: The seven lysine side chains are shown in orange; http://en.wikipedia.org/
Ubiquitin is a small protein that exists in all eukaryotic cells. It performs its myriad functions through conjugation to a large range of target proteins. A variety of different modifications can occur. The ubiquitin protein itself consists of 76 amino acids and has a molecular mass of about 8.5 kDa. Key features include its internal 7 Lys residues and a C-terminal tail with two Glycine residues. It is highly conserved among eukaryotic species: Human and yeast ubiquitin share 96% sequence identity. And still, protein modification by covalent conjugation of the ubiquitin molecule is one of the most dynamic posttranslational modifications studied in terms of biochemistry and cell physiology. Ubiquitination plays a central regulatory role in number of eukaryotic cellular processes such as receptor endocytosis, growth-factor signaling, cell-cycle control, transcription, DNA repair, gene silencing, and stress response.
NH3^+MQIFVKTLTGKTITLEVEPSDTIENVKAKQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG-COO^-
Ubiquitination is an enzymatic, protein post-translational modification (PTM) process in which the carboxylic acid of the terminal glycine from the di-glycine motif in the activated ubiquitin forms an amide bond to the epsilon amine group of the lysine in the target protein.
- This process of degradation is inhibited under anaerobic conditions. The process requires ATP energy and it is lysosome independent.
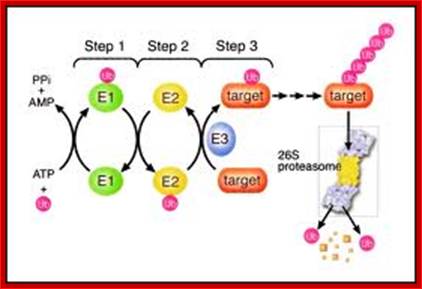
The ubiquitin-proteasome pathway. Proteins marked with a polyubiquitin chain by the E1-E2-E3 enzymatic cascade are targeted for degradation by the proteasome. A ubiquitin-activating enzyme (E1) binds ubiquitin in an adenosine triphosphate (ATP) –dependent step. Ubiquitin is then transferred to a ubiquitin-conjugating enzyme (E2). A ubiquitin ligase (E3) helps transfer ubiquitin to the target substrate. PPI, pyrophosphate; AMP, adenosine monophosphate; Ub, ubiquitin. (Figure and legend adapted30 and used by permission from Elsevier.); The Ubiquitin-Proteasome Pathway; http://jco.ascopubs.org/
Ubiquitin + HS-E1 + ATP----> *Ubiquitin—C (=O)~S-E1 = ADP + P
*Ubiqitin-C(=O)-S-E1 + E2-SH --->*Ubiquitin-S~E2 + E1
*Ubiquitin-S~E2 + E3-SH Protein-Lysine à Protein-Lysine-*Ubiquitin + E3 ligase
www.biocompare.com
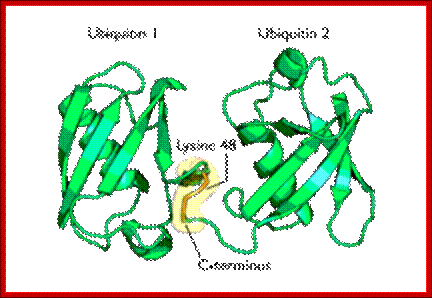
Diagram of lysine 48-linked diubiquitin: The linkage between the two ubiquitin chains is shown in orange. Such kind of linkages is possible with Lysine 63
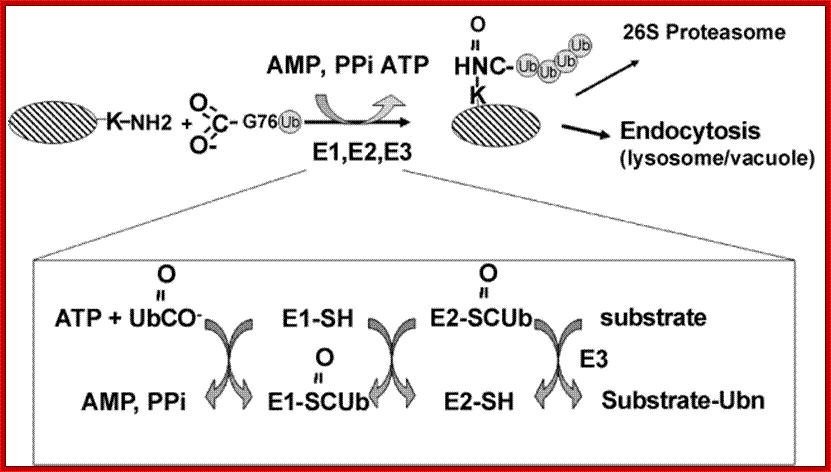
Ubiquitination reaction. The protein substrate is ubiquitinated in a reaction involving three types of ubiquitinating enzymes: the ubiquitin activating protein E1, an ubiquitin carrier protein E2, and an ubiquitin-protein ligase E3. Following addition of a single ubiquitin molecule to a protein substrate (monoubiquitination), further ubiquitin molecules can be added to the first, yielding a polyubiquitin chain. The fate of the protein depends on the type of ubiquitin chain formed on the protein substrate; www.omicsonline.org Trafina Jadhav and Marie W. Wooten.
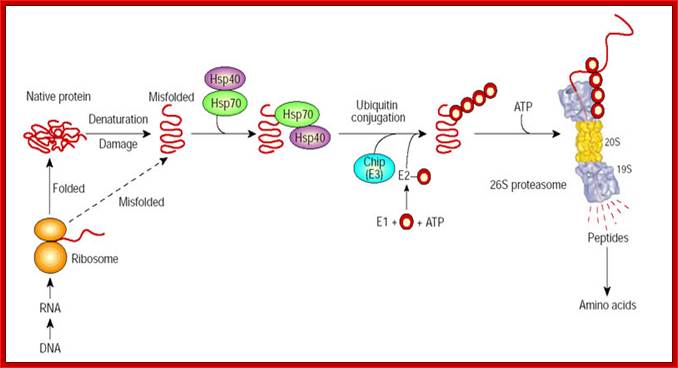
Molecular chaperones may function in protein folding and in the degradation of misfolded species. By associating with exposed hydrophobic domains, chaperones Hsp70/40 promote the folding of newly synthesized proteins and favours their refolding. Alternatively, they can facilitate the recognition of abnormal proteins, leading to their ubiquitylation by CHIP, the E3, and their degradation by the 26S proteasome. The red circles represent ubiquitin. Alfred L. Goldberg; http://www.nature.com/
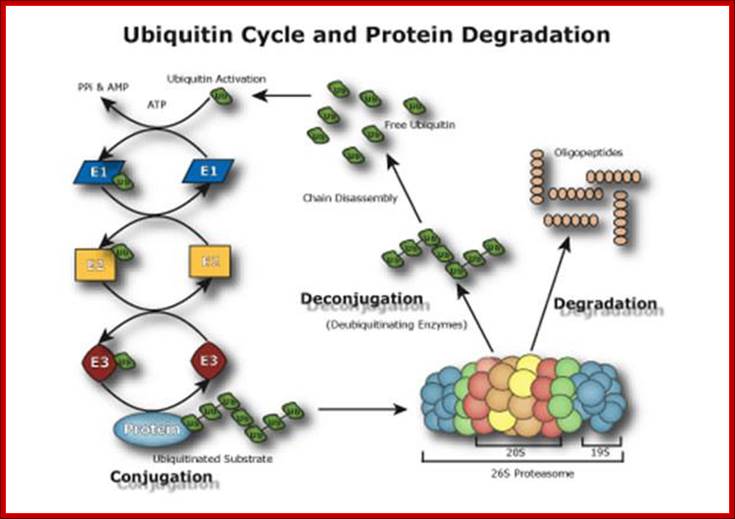
Ubiquitin Cycle Pathway (UPP); The Ubiquitin Proteasome Pathway (UPP) is the principal mechanism for protein catabolism in the mammalian cytosol and nucleus. The highly regulated UPP affects a wide variety of cellular processes and substrates and defects in the system can result in the pathogenesis of several important human diseases. The central role of the UPP in biology has been recognized with the Nobel Prize for Chemistry which was awarded to Avram Hershko, Aaron Ciechanover and Irwin Rose in 2004. The UPP is central to the regulation of almost all cellular processes including: http://www.bostonbiochem.com/about/ubiquitin-proteasome-pathway-upp
Ubiquitin protein is ubiquitous in cells, monomeric and contains 76 amino acids. Its amino acid sequences are highly conserved among the species like humans, Toads and Drosophila. The difference between yeast’s ubiquitin and humans is a difference of three amino acids.
- Covalent addition of ubiquitins to selected proteins is called ubiquitination; this marks proteins, for selected proteolysis.
- Ubiquitins at COO^- terminal end have glycine residue.
- Conjugation involves attachment of C-terminal glycine of ubiquitin (Ub) to the ε-amino group in lysine residues of the targeted protein. The conserved conjugation reaction is achieved by sequential actions of three enzymes
- There are ubiquitin-activating enzymes called E1, E2 and E3.
- Ubiquitins are first conjugated to E1 enzyme, which has SH group; this thioester linking of carboxyl group with SH of E1 requires ATP energy.
Ubiquitin + HS-E1 + ATP----> Ubiquitin—C (=O)~S-E1 = ADP + P
- Then the second enzyme, E2-SH, which is a protein conjugating enzyme, transfers ubiquitin’s C=O group to its own SH group to form thioester bond.
- There are different kinds of EF2.
- The core of EF2s is about 150 amino acids. These proteins just vary from one another at their N or C terminal regions. Most of the EF-2s from a variety of sources show amino acid homology by 42%. They are structurally super imposable. Many a times EF-2 s by themselves, can conjugate ubiquitins to target proteins.
Ubiqitin-C(=O)-S-E1 + E2-SH --->Ubiquitin-S~E2 + E1
- The E3-SH are 180kd proteins they act as ligases, which performs conjugation between the NH2 group of Lysine (at epsilon position) and –C=O group of the ubiquitin to form ubiquitin-C (=O)-NH-, which is an Isopeptide bond. In the process the E2-SH is freed.
- Ubiquitin-S~E2 + E3-SH Protein-Lysine à Protein-Lysine-Ubiquitin + E3 ligase.
- By this mechanism, all those lysines found in the condemned proteins are ubiquitinated. In addition, each of the ubiquitins is further ubiquitinated to produce tandem repeats of ubiquitins (multiubiquitins).
- Ubiquitins can be ubiquitinated so as to identify the protein easily.
- Such ubiquitinated proteins provide landmarks for recognition and they are fed to proteolytic enzyme complex called Proteasomes.
“The process of marking a protein with ubiquitin (ubiquitylation or ubiquitination) consists of a series of steps: (Repeat)
- Activation of ubiquitin: Ubiquitin is activated in a two-step reaction by an E1 ubiquitin-activating enzyme in a process requiring ATP as an energy source. The initial step involves production of a ubiquitin-adenylate intermediate. The second step transfers ubiquitin to the E1 active site cysteine residue, with release of AMP. This step results in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryls group.
- Transfer of ubiquitin from E1 to the active site cysteine of a ubiquitin-conjugating enzyme E2 via a trans-(thio) esterification reaction. Mammalian genomes contain 30-40 UBCs.
- The final step of the ubiquitylation cascade creates an isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin. In general, this step requires the activity of one of the hundreds of E3 ubiquitin-protein ligases (often termed simply ubiquitin ligase). E3 enzymes function as the substrate recognition modules of the system and are capable of interaction with both E2 and substrate.”
In the ubiquitination cascade E1 can bind with dozens of E2s which can bind with hundreds of E3s in a hierarchical way. Other ubiquitin-like proteins (ULPs) are also modified via the E1–E2–E3 cascade.
The most studied polyubiquitin chains – lysine 48-linked - target proteins for destruction, a process known as proteolysis. At least four ubiquitin molecules must be attached to lysine residues on the condemned protein in order for it to be recognized by the 26S-proteasome. The ubiquitin molecule provide lysine sites for more ubiquitination, thus poly-ubiquitination is possible. Ubiquitination is not just linked to only to lysine residues they can be linked to serine, threonine and so on. Deubiquitination is also possible.
The proteasome is a complex, barrel-shaped structure with two chambers, within which proteolysis occurs. Proteins are rapidly degraded into small peptides (usually 3-24 amino acid residues in length). Ubiquitin molecules are cleaved off the protein immediately prior to destruction and are recycled for further use. Although the majority of proteasomal substrates are ubiquitinated, there are examples of non-ubiquitinated proteins targeted to the proteasome.
The ubiquitination system functions in a wide variety of cellular processes, including. Ubiquitination is also used for -Antigen processing, Apoptosis, Biogenesis of organelles, Cell cycle and division, DNA transcription and repair, Differentiation and development, Immune response and inflammation, Neural and muscular degeneration, Morphogenesis of neural networks, Modulation of cell surface receptors, ion channels and the secretory pathway, Response to stress and extracellular modulators, Ribosome biogenesis, Viral infection and others
Deubiquitinating enzyme; (From Wikipedia, the free encyclopedia);
Deubiquitinating enzymes (DUBs) are a large group of proteases (more than 60 known) that regulate ubiquitin-dependent metabolic pathways by cleaving ubiquitin-protein bonds. DUBs are also commonly referred to as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolyases, ubiquitin isopeptidases, or DUbs. The human genome encodes nearly 100 DUBs with specificity for ubiquitin in five gene families. Potentially, DUBs may act as negative and positive regulators of the ubiquitin system. In addition to ubiquitin recycling, they are involved in processing of ubiquitin precursors, in proofreading of protein ubiquitination and in disassembly of inhibitory ubiquitin chains. DUBs can be classified into two main classes: cysteine proteases and metalloproteases.
DUBs role in the ubiquitin pathway:
DUBs play several roles in the ubiquitin pathway. First, DUBs carry out activation of the ubiquitin proproteins, probably cotranslationally. Second, DUBs recycle ubiquitin that may have been accidentally trapped by the reaction of small cellular nucleophiles with the thiol ester intermediates involved in the ubiquitination of proteins. Third, DUBs reverse the ubiquitination or ubiquitin-like modification of target proteins. Finally, DUBs are also responsible for the regeneration of monoubiquitin from unanchored polyubiquitin, i.e., free polyubiquitin that is synthesized de novo by the conjugating machinery or that has been released from target proteins by other DUBs.
SCF proteins and their role:
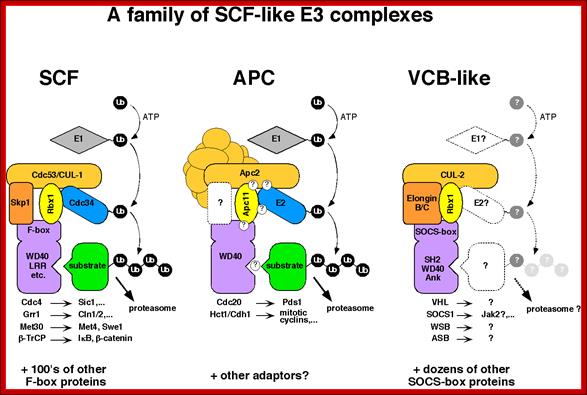
www.lightinghomes.ne
The above diagram shows few complexes such as SCF (SKp1, cdc53/Cullin and Fox protein complex), APC (Anaphase promoting complex) and VCB like (VHl, Socs, WSb, and Asb complex). They are also involved in conjugating Ubiquitin to target proteins using E1, E2 and E3 ligase. They lead the target proteins to degradation.
These individual complexes act as E3 ligase complexes, where activated ubiquitins or ubiquitin like proteins are added to target proteins. ScF acts at G1 and S phase transition, and it is responsible for the degradation of Sic1 (stoichiometric inhibitor of Cdk1-Clb B-type cyclins) and cln1/2 and cdc28, so facilitates cell cycle go into S-phase. Similarly APC complex called as cyclosome acts on Cyclin B/Cdk2 (MPFs) to remove them. It is this action leads to the entry and completion of Anaphase leading to telophase and mitosis stages. VCB like complex also act as E3 ligase but the marking proteins are different; perhaps they are Ubiquitin like Proteins.
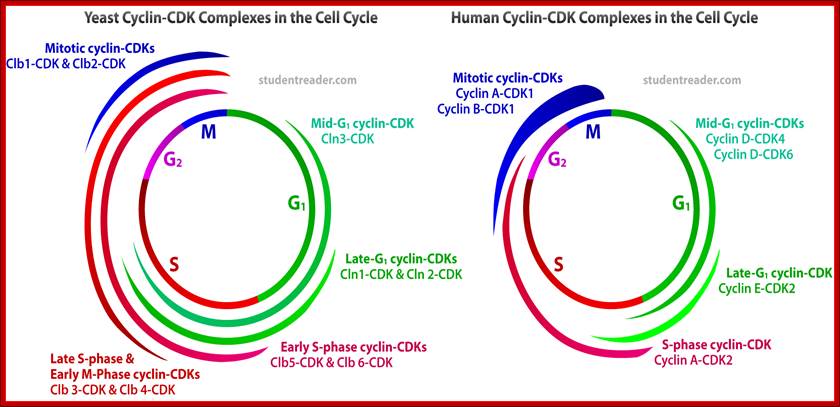
An obvious advantage of proteolysis for controlling passage through these critical points in the cell cycle is that protein degradation is an irreversible process, ensuring that cells proceed irreversibly in one direction through the cycle. http://studentreader.com/
Proteasome complex:
- Proteasome is a complex of proteases organized into a barrel shaped structure, made up of 28 subunits; its total Mol.wt is 2000kDa. It consists of at least five different peptidase functions. They cleave after basic, hydrophobic or hydrophilic amino acid residues.
- Ubiquitylated proteins are fed into proteasome, assisted by chaperones, and the proteins are degraded in ATP dependent manner.
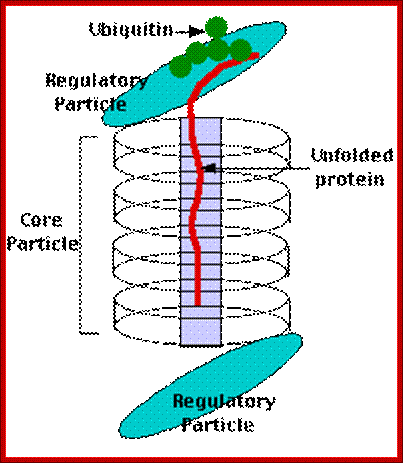
Biology pages; users.rcn.com/ jkimbeall.ma.ultranet /
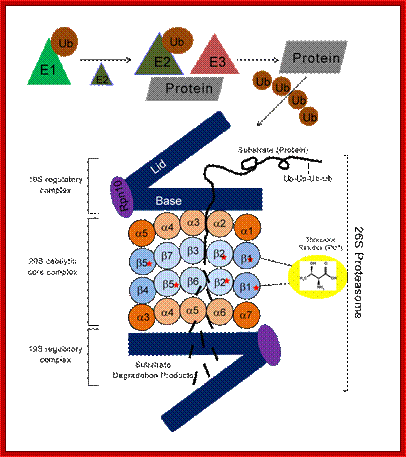
Samuel Troy Pellom Jr and Anil Shankar .;http://omicsonline.org/

; Nodel for human proteasome; Red subunits were used in pull-down assays and analyzed with mass spectrometry. Yellow and red subunits were used in protein:protein mapping experiments. Green subunits are found in the 19S regulatory particle. Orange subunits are found in the 20S core. Models were based on yeast proteasome and human mapping data.
;Brad Hook and Trista Schagat;http://www.promega.es/
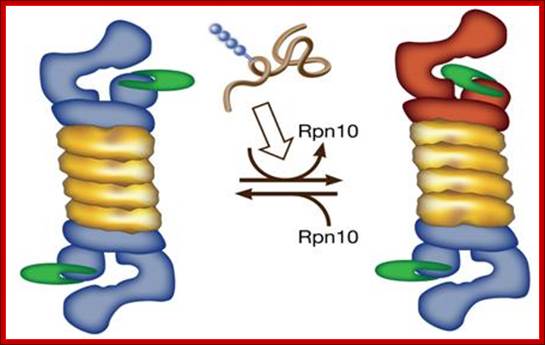
A possible model for polyubiquitinated substrate-dependent conformational transition in the 19S proteasome regulatory complex, presumably, mediated through disassociation of S5a/ Rpn10. Architecture of 19S proteasome; http://wws.weizmann.ac.il/

- This process of degradation is inhibited under anaerobic conditions and requires ATP energy and it is lysosome independent.
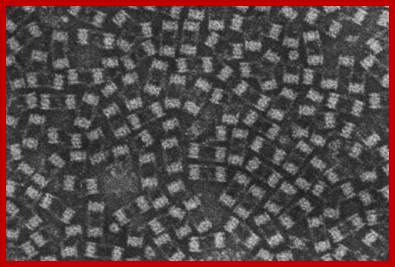
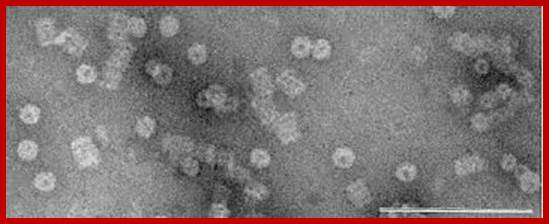
Protreasomes in Archaea; Julie A. Maupin-Furlow, et al ;http://www.bioscience.org/
Crystallized Proteasomes in Archaea; Julie A. Maupin-Furlow, http://www.bioscience.org/
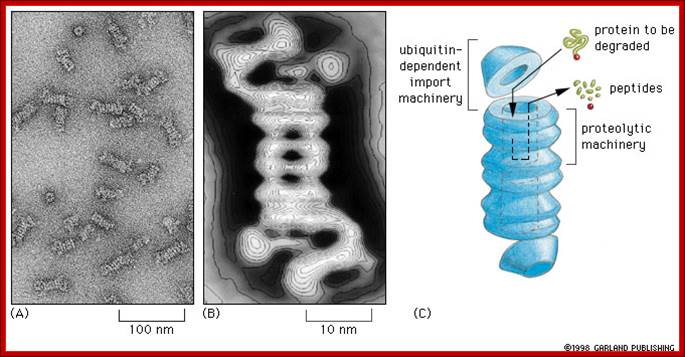
Proteasomes degrade unwanted proteins in the cytosol compartment. (A) EM of proteasomes (B) Computer enhanced image (C) Artist's conception of how it works. The red ball attached to the protein to be degraded (green) represents the ubiquitin peptide that is attached to the unwanted protein to tag it for destruction.;http://www.zoology.ubc.ca/

http://flipper.diff.org
Proteasome consists of barrel shaped structure with orifices at both ends. At least five different peptidases functions are located at the inner face of the barrel shaped structure.
The
26S proteasome imaged by electron microscopy. Right figure; the model of
the 26S proteasome showing the architecture of the subunits. Molecular
organization of the 26S proteasome. (Left): Averaged image based on electron
micrographs of the complex of the 26S proteasome from rat. The α and
β rings of the 20S proteasome are indicated. Photograph kindly provided by
W. Baumeister. (Right): Schematic drawing of the subunit structure. Ub,
ubiquitin;
(Ub-activating), E2 (Un-conjugating), and E3
(Ub-ligating) enzymes; CP, core particle; RP, regulatory particle; Rpn, RP
non-ATPase; Rpt, RP triple ATPase; Ub-R, Lib receptor (poly-Ub binding subunit).
The diagram shows ubiquitination and deubiquitination (DUB) cycle. Which proteins are degraded by proteasome? Stephan Mann; http://flipper.diff.org/
Left fig-Active-site directed probes to report enzymatic action in the ubiquitin proteasome system; fig on right side shows the inner surface of the chaperonins.
On the inner face of protein subunits have protein binding and cleavage activity, thus the the target proteins that enter are subjected to cleavage and the smaller peptides or even amino acids are released from the other side of the the proteasome barrel. It is interesting to compare the overall structure of proteasome and protein modification barrels looks almost similar. Active-site directed probes to report enzymatic action in the ubiquitin proteasome system; ;Huib Ovaa; ;http://www.nature.com/ GroEL and GroES innersurface where proteins are processed to proper folding; tem Polypeptidhttp://www.chemgapedia.de/
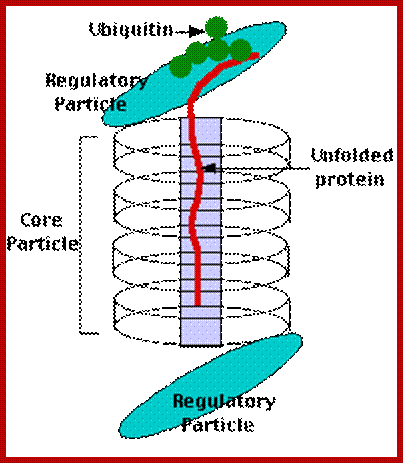
http://flipper.diff.org
- These huge protein complexes are found both in the nucleus and cytoplasm.
- Proteasome complex is made up of 4 rings of protein subunits called alpha and beta.
- Each ring consists of 7 subunits. They are laid one over the other. The top and the bottom rings are made up of alpha units and the middle two rings contain beta subunits.
- It is at the inner surface of the beta ring one finds protease domains.
- They appear as barrel shaped structures of 450 x 190 Å, with a hallow in the center. It is into this hallow region the ubiquitinated proteins are fed and the proteins are degraded in ATP dependent manner.
- In addition to this barrel shaped structure one finds another 19s protein complex at both ends of the barrel protein complex. This almost acts like guard or gate and feeds the ubiquitinated proteins into the barrel in ATP dependent manner. This cap and barrel shaped structure is 26s complex.
The Core Particle (CP)
- The core particle is made of 2 copies of each of 14 different proteins.
- These are assembled in groups of 7 forming a ring.
- The 4 rings are stacked on each other (like 4 donuts)
The Regulatory Particle (RP)
- There are two identical RPs, one at each end of the core particle.
- Each is made of 18 different proteins (none of them the same as those in the CP).
- 6 of these are ATPases.
- Some of the subunits have sites that recognize the small protein ubiquitin.
Ubiquitylation in the ERAD Pathway: Frederik Eisele, Antje Schäfer and Dieter H. Wolf
Ubiquitylation is a protein modification mechanism, which is found in a multitude of cellular processes like DNA repair and replication, cell signaling, intracellular trafficking and also, very prominently, in selective protein degradation. One specific protein degradation event in the cell concerns the elimination of misfolded proteins to prevent disastrous malfunctioning of cellular pathways. The most complex of these ubiquitylation dependent elimination pathways of misfolded proteins is associated with the endoplasmic reticulum (ER). Proteins, which enter the endoplasmic reticulum for secretion, are folded in this organelle and transported to their site of action. A rigid protein quality control check retains proteins in the endoplasmic reticulum, which fail to fold properly and sends them back to the cytosol for elimination by the proteasome. This requires crossing of the misfolded protein of the endoplasmic reticulum membrane and polyubiquitylation in the cytosol by the ubiquitin-activating, ubiquitin-conjugating and ubiquitin-ligating enzyme machinery

https://beyondthedish.wordpress.com
Ubiquitylation is required for different steps of the ER associated degradation process (ERAD). It facilitates efficient extraction of the ubiquitylated misfolded proteins from and out of the ER membrane by the Cdc48‑Ufd1‑Npl4 complex and thereby triggers their retro translocation to the cytosol. In addition, the modification with ubiquitin chains guarantees guidance, recognition and binding of the misfolded proteins to the proteasome in the cytosol for efficient degradation
Bacterial Proteasomes:
Bacterial Proteasomes are made up of two rings of proteins. Each ring consists of 6 subunits each. The upper one is called alpha and the lower one is called beta ring. Eubacterial proteasome structural elements are like forerunners of eukaryotic proteasome subunits.
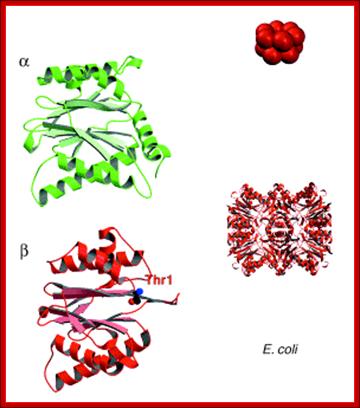
Presently prokaryotic Proteasome consists of ClpXp protease. The upper figure shows two rings of six protein subunits; the bottom figure shows a barrel shaped structure similar to that of eukaryotic proteasome.
A new way to identify bacterial degradation signals: ; A proteasome of bacterial origin, They contain ClpXp proteases and organized into single barrel like structure into which abnormal proteins are fed and digested; Cecile M. Pickart & Robert E. Cohen http://www.zoology.ubc.ca; http://www.nature.com
In the past, degradation signals have been identified through structure–function analysis of known natural substrates (Box 1). However, in a recent experiment, Flynn et al.62 used a protease-dead version of ClpXP to capture intact substrates in vivo (see figure; see also Ref. 60). After capture, intact substrates were isolated by ClpXP affinity purification, identified and subjected to sequence analysis to identify common motifs. This experiment identified many new substrates, and comparing their sequences identified several new bacterial degradation signals. Due to the essentiality of the 26S proteasome, this approach is not directly feasible for this enzyme. However, using proteomics techniques, targets of ubiquitin conjugation can be identified on a proteome-wide scale98, and most of these proteins are substrates of the proteasome. Proteasomes and their kin: proteases in the machine age Nature Reviews- Molecular Biology.
It has been suggested, that during expression of several genes, ubiquitins are targeted to H2A histones, thus the nucleosomal free DNA is formed, which is used for the assembly of transcriptional factors and the required enzymes bind for gene expression. It is also viewed that ubiquitin dependent proteolysis requires tRNA linking?
In the past, degradation signals have been identified through structure–function analysis of known natural substrates (Box 1). However, in a recent experiment, Flynn et al. used a protease-dead version of ClpXP to capture intact substrates in vivo (see figure; see also Ref. 60). After capture, intact substrates were isolated by ClpXP affinity purification, identified and subjected to sequence analysis to identify common motifs. This experiment identified many new substrates, and comparing their sequences identified several new bacterial degradation signals. Due to the essentiality of the 26S proteasome, this approach is not directly feasible for this enzyme. However, using proteomics techniques, targets of ubiquitin conjugation can be identified on a proteome-wide scale98, and most of these proteins are substrates of the proteasome; Cecile M. Pickart & Robert E. Cohen
Sumoylation:
Small Ubiquitin-like Modifier or SUMO proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. SUMOylation is a post-translational modification involved in various cellular processes, such as regulation of chromatin structure, DNA repair, nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress and progression through the cell cycle.
Contrary to the long-standing assumption that SUMO has no role in proteolytic targeting and rather acts as an antagonist of ubiquitin in some cases, it has recently been discovered that sumoylation itself can function as a secondary signal mediating ubiquitin-dependent degradation by the proteasome. The discovery of a novel family of RING finger ubiquitin ligases bearing SUMO interaction motifs implicated the ubiquitin system in the control of SUMO modified proteins. SUMO modification as a signal for degradation is conserved in eukaryotes and ubiquitin ligases that specifically recognize SUMO-modified proteins have been discovered in species ranging from yeasts to humans. This review summarizes what is known about these ligases and their role in controlling sumoylated proteins; Maria Miteva et al MadamCurie Bioscience Database 2010.

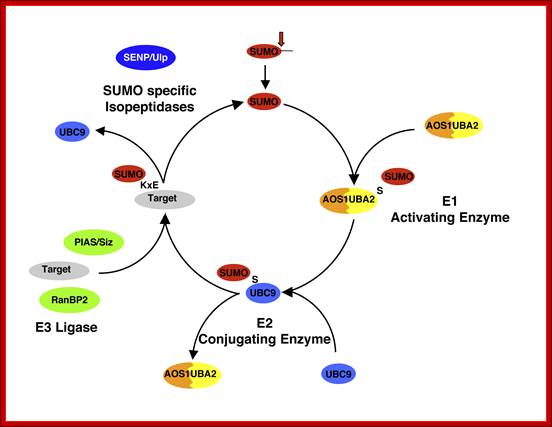
Figure.Comparison of ubiquitin and SUMO conjugation systems. Ubiquitin (Ub) and SUMO conjugation involves the activities of related enzymes. A) After processing of its precursor forms by deubiquitylating enzymes (Dub), Ub is activated by Ub-activating enzyme (E1) und subsequently conjugated to substrates by complexes of Ub-conjugating enzymes (E2) and Ub ligases (E3). Attachment of Ub to Ub leads to the formation of substrate attached chains. Ub isopeptidases (Dub) can regenerate free Ub from substrates. B) The analogous enzymes as shown in A) are shown for the SUMO system. Here the activating enzyme is composed of two subunits (Uba2 and Aos1) and only a single conjugating enzyme (Ubc9) is used. Desumoylating enzymes are Ulp1 and Ulp2 in budding yeast and several SENPs in mammals; Maria Miteva et al MadamCurie Bioscience Database 2010. www.keywordsking.com
In recent years, scientists have discovered ubiquitin like molecules (UBLs). One of them is SUMO- means small ubiquitin like modifiers. They are more or less homologous to ubiquitins, but SUMO proteins are 100 amino acids long.
Small Ubiquitin-like Modifier (or SUMO) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle (Wikipedia). In contrast to ubiquitin, SUMO is not used to tag proteins for degradation. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine on the target protein.
Structure:
Sumo1 Protein; WIKIhttps://commons.wikimedia.org/
SUMO proteins are small; most are around 100 amino acids in length and 12 kDa in mass. The exact length and mass varies between SUMO family members and depends on which organism the protein comes from. Although SUMO has very little sequence identity with ubiquitin at the amino acid level, it has a nearly identical structural fold.
The structure of human SUMO1 is depicted above. It shows SUMO1 as a globular protein with both ends of the amino acid chain (shown in red and blue) sticking out of the protein's centre. The spherical core consists of an alpha helix and a beta sheet. The diagrams shown are based on an NMR analysis of the protein in solution
Sumo activating enzymes SAE1 and SAE2 is transferred to Sumo. ATP is required for this conjugation. Sumo conjugating enzyme Ubc9-SH transfers to Sumo-S-Sae 1 and 2 and releases SAE1 and SAE2. Suo-S-Ubc9 is transferred to target protein; thus protein for destruction is marked.

Sumoylation steps; Pathway for Sumo conjugation. Sumo is a -100 residue protein that is conjugated to a number of proteins via an isopeptide linkage. The conjugation pathway is completely analysis to the ubiquitylation pathway. While ubiquitylation leads to protein degradation sumoylation has diverse consequences on protein behavior. http://www.biochemistry.ucla.edu/biochem
Prediction of SUMO attachment
Most SUMO-modified proteins contain the tetra peptide consensus motif Ψ-K-x-D/E where Ψ is a hydrophobic residue, K is the lysine conjugated to SUMO, x is any amino acid (aa), D or E is an acidic residue. Substrate specificity appears to be derived directly from Ubc9 and the respective substrate motif. SUMOplot is online free access software developed to predict the probability for the SUMO consensus sequence (SUMO-CS) to be engaged in SUMO attachment. The SUMO plot score system is based on two criteria: 1) direct amino acid match to the SUMO-CS observed and shown to bind Ubc9, and 2) substitution of the consensus amino acid residues with amino acid residues exhibiting similar hydrophobicity. SUMOplot has been used in the past to predict Ubc9 dependent sites. Alternative prediction engines such as SUMOsp are also available.
SUMO Attachment
SUMO attachment to its target is similar to that of ubiquitin (as it is for the other ubiquitin-like proteins such as NEDD 8. A C-terminal peptide is cleaved from SUMO by a protease (in human the SENP proteases or Ulp1 in yeast) using ATP and di-glycine motif. SUMO then becomes bound to an E1 enzyme (SUMO Activating Enzyme (SAE)) which is a heterodimer. It is then passed to an E2 which is a conjugating enzyme (Ubc9). Finally, one of a small number of E3 ligating proteins attaches it to the protein. In yeast, there are four SUMO E3 proteins, Cst9, Mms21, Siz1 and Siz2. While in ubiquitination an E3 is essential to add ubiquitin to its target, evidence suggests that the E2 is sufficient in Sumoylation as long as the consensus sequence is present. It is thought that the E3 ligase promotes the efficiency of sumoylation and in some cases has been shown to direct SUMO conjugation onto non-consensus motifs. E3 enzymes can be largely classed into PIAS proteins, such as Mms21 (a member of the Smc5/6 complex) and Pias-gamma and HECT proteins. Some E3's such as RanBP2 however are neither. Recent evidence has shown that PIAS-gamma is required for the sumoylation of the transcription factor yy1 but it is independent of the zinc-RING finger (identified as the functional domain of the E3 ligases). SUMOylation is reversible and is removed from targets by specific SUMO proteases in an ATP dependent manner. In budding yeast, the Ulp1 SUMO protease is found bound at the nuclear pore, whereas Ulp2 is nucleoplasmic. The distinct sub nuclear localization of deSUMOylating enzymes is conserved in higher eukaryotes.
The pathway of SUMO conjugation/deconjugation: SUMO is synthesised as a precursor and C-terminally processed (arrowhead). Subsequently, the conjugation to proteins involves the E1 SUMO activating enzyme (AOS1/UBA2) and an E2 conjugating enzyme (Ubc9) that form thioesters (S) with the modifier. E3-like factors, like PIAS and RanBP2, stimulate the attachment to lysine residues within a target protein. The cleavage of SUMO from its target proteins, termed “de-sumoylation”, is catalysed by members of the SENP isopeptidase family. In humans six members of this family were identified. www.loosenessofassociation.com
DeSUMOylating enzymes-SENPs: (M Drag, GS Salvesen)
Modification of proteins by ubiquitin and SUMO (small ubiquitin-like modifiers) is a dynamic and reversible process. Similar to the ubiquitin pathway, where the action of deubiquitinating enzymes removes ubiquitin from ubiquitin-adducts, SUMO is also removed intact from its substrates by proteases belonging to the sentrin-specific proteases (SENPs) family. In addition to their isopeptidase activity, SENPs also execute another essential function as endopeptidases by removing the short C-terminal extension from immature SUMOs. The defining characteristics of SENPs are their predicted conserved molecular scaffold-defined as members of peptidase Clan CE, conserved catalytic mechanism, and their reported activity on SUMO or Nedd8 conjugated proteins (or the respective precursors). We discuss recent progress on the human SENPs and their substrates. IUBMB IUBMB Life, 2008.
SUMOylation and De-SUMOylation: Wrestling with Life's Processes*
By Edward T. H. Yeh.
The small ubiquitin-like modifier (SUMO) is a ubiquitin-like protein that covalently modifies a large number of cellular proteins. SUMO modification has emerged as an important regulatory mechanism for protein function and localization. SUMOylation is a dynamic process that is mediated by activating (E1), conjugating (E2), and ligating (E3) enzymes and readily reversed by a family of ubiquitin-like protein-specific proteases (Ulp) in yeast and sentrin/SUMO-specific proteases (SENP) in human. This note will focus on the de-SUMOylating enzymes with special attention to their biological function.
Many biochemical pathways are reversible to create an on and off state that is essential for biological regulation. A reversible system allows for quick termination of a biological response that has to be precisely controlled. The SUMO2 modification pathway is an example of a reversible system that is controlled by a series of on and off enzymes. In contrast to the much more complex ubiquitin pathway, SUMOylation utilizes only a single conjugating enzyme, Ubc9, and a limited number of ligases. This simplicity also manifests in the off step because there are only two SUMO-deconjugating enzymes in yeast and six in human. One may assume that these limited numbers of on and off enzymes would sufficient to regulate only be a small number of substrates and biological pathways. However, the number of SUMO substrates continues to expand, and the varieties of systems that are known to be regulated by SUMO also proliferate quickly. The de-SUMOylation pathways provide insights into how these limited numbers of proteases are able to regulate a diverse array of biological responses.
SUMOylation and de-SUMOylation: SUMOylation is a dynamic process that is mediated by activating (E1), conjugating (E2), and ligating (E3) enzymes and readily reversed by the SENP family in human. SUMOylation and de-SUMOylation regulate a diverse spectrum of biological responses, from transcription, cell division, and signal transduction to carcinogenesis and viral replication.
It has recently been discovered that SUMOylation itself can function as a secondary signal mediating ubiquitin‑dependent degradation by the proteasome. The discovery of a novel family of RING finger ubiquitin ligases bearing SUMO interaction motifs implicated the ubiquitin system in the control of SUMO modified proteins. SUMO modification as a signal for degradation is conserved in eukaryotes and ubiquitin ligases that specifically recognize SUMO‑modified proteins have been discovered in species ranging from yeasts to humans. This review summarizes what is known about these ligases and their role in controlling sumoylated proteins; Maria Miteva, Kirstin Keusekotten, Kay Hofmann, Gerrit J.K. Praefcke and R. Jürgen Dohmen
Although the post-translational modification of proteins with small ubiquitin-like modifier (SUMO) has a role in many biological processes, it was thought that SUMO, unlike ubiquitin, does not target proteins for degradation. However, these views need to be revised, as recent findings in yeast and human cells indicate that SUMO can act as a signal for the recruitment of E3 ubiquitin ligases, which leads to the ubiquitylation and degradation of the modified protein. This is An additional role for SUMO in ubiquitin-mediated proteolysis;
Marie-Claude Geoffroy1 & Ronald T. Hay1

The effect of specific kinase inhibitors that can block small ubiquitin-like modifier (SUMO) modification of promyelocytic leukaemia protein (PML; also known as TRIM19), suggests that phosphorylation of PML is a key step in its arsenic-induced degradation44, 53. Arsenic-induced phosphorylation of PML might occur through a mitogen-activated protein kinase (MAPK) pathway, in particular through the activation of ERKs (extracellular signal-regulated kinases), JNKs (JUN N-terminal kinases) and p38, resulting in increased SUMO modification and apoptotic activity of PML54, 55, 56. It has also been shown that arsenic induces the DNA damage response kinase ATR (ataxia telangiectasia and Rad3-related protein), which seems to function upstream of PML in a signal transduction pathway that also activates cell cycle checkpoint kinase 2 (CHK2) and p53, leading to arsenic-induced apoptosis57. Phosphorylation, in turn, activates or recruits an E3 SUMO ligase that generates a SUMO chain on PML in the presence of an E1 SUMO-activating enzyme (SAE1 and SAE2) and an E2 SUMO-conjugating enzyme (ubiquitin carrier 9 (UBC9); also known as UBE2I). This SUMO chain then engages the SIM domains of RING finger protein 4 (RNF4; also known as SNURF), which uses its E3 ubiquitin ligase activity to add polyubiquitin chains to the SUMO polymer and PML, thus targeting them for ubiquitin-mediated degradation by the 26S proteasome. This process is highly regulated in vivo by the SUMO-specific proteases SENP1–3 and SENP5–7, which process SUMO precursors and deconjugate SUMO from the substrate6. Additionally, deubiquitylating enzymes (DUBs) can also deconjugate ubiquitin from modified proteins to limit protein degradation. Nature Reviews-molecular biology 2009;Model for RNF4-dependent degradation; Marie-Claude Geoffroy & Ronald T. Hay
Sumoylation as a Signal for Polyubiquitylation and Proteasomal Degradation:
The small ubiquitin‑related modifier (SUMO) is a versatile cellular tool to modulate a protein’s function. SUMO modification is a reversible process analogous to ubiquitylation. The consecutive actions of E1, E2 and E3 enzymes catalyze the attachment of SUMO to target proteins, while deconjugation is promoted by SUMO specific proteases. Contrary to the long‑standing assumption that SUMO has no role in proteolytic targeting and rather acts as an antagonist of ubiquitin in some cases, it has recently been discovered that sumoylation itself can function as a secondary signal mediating ubiquitin‑dependent degradation by the proteasome. The discovery of a novel family of RING finger ubiquitin ligases bearing SUMO interaction motifs implicated the ubiquitin system in the control of SUMO modified proteins. SUMO modification as a signal for degradation is conserved in eukaryotes and ubiquitin ligases that specifically recognize SUMO‑modified proteins have been discovered in species ranging from yeasts to humans. This summarizes what is known about these ligases and their role in controlling sumoylated proteins; Maria Miteva, Kirstin Keusekotten, Slx5-Slx8 and its orthologs are proposed to target SUMO conjugates for ubiquitin-mediated proteolysis, but the only in vivo substrate identified to date is mammalian PML, and the physiological importance of SUMO-targeted ubiquitylation remains largely unknown, Zheng Wang and gregoru Prelich

Figure. STUbL Pathway. (I-II) After stimulus, target X becomes sumoylation competent (e.g. phosphorylated) and is SUMO conjugated. (III) Following execution of the SUMO conjugated target function, STUbL-mediated ubiquitination, perhaps coupled to desumoylation by Ulp1/2, promotes target proteasomal target degradation (IV). SUMO recycling may also occur at the proteasome as for ubiquitin; http://www.scripps.edu/boddy.
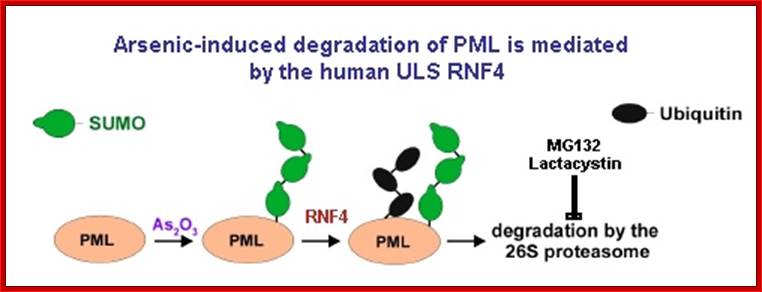
Uzunova et al., 2007; Weisshaar et al., 2008; Sumo conjugates with ubiqui5tin that leads to proteasome mediated degradation