Cellular Signal Transduction I:
Cells and Signals:
Introduction:
Organisms’ prokaryotes or eukaryote plants and animals are always surrounded by environment and acted upon by environmental components, such as air, water, wind, light, temperature, chemicals, microbes and others. Intracellular organelles too live with fluid cytoplasm; many chemicals generate which act on cells for certain responses ex. during developmental processes.
Environmental signals impinge upon the surface of the body and body is made up cells; and the body responds. Visual signals are many, one example such as a tiger or a snake can cause fear instaneously, the body temperature increases within minutes, greater amount of energy generated, which can be used to fight or to run (flight) and save her or his life (selfish-inbuilt).
The body of an organism is covered by cell wall in the case of plants and bacteria and outer cellular layers such as epidermal skin or epithelium in the digestive tract, urogenital tracts, eye and ear. Cells are endowed with surface structures to receive any kind of environmental or chemical or both signals. Cells contain on their surface a variety of signal receivers or sensors called receptors, specific to specific cell type and specific to specific signals. The number and kinds of receptors vary from one cell type to the other. All put together any cell will contains at least few thousands of them on the surface of each cell. Receptors are also found within the cytoplasm (intracellular) or in extra cellular fluids and matrixes.
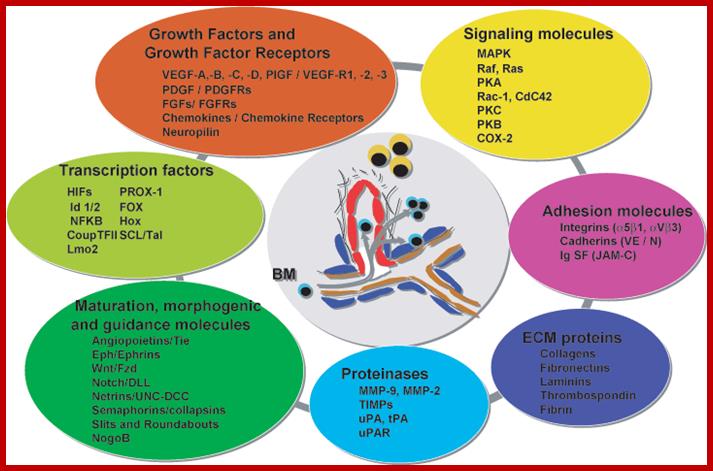
Cells in the body or tissue live in a community and they communicate with each other either by direct contact or through chemical signals that activate or depress cellular activities. Those that activate are called Agonists and those that act against are called Antagonists. In a cellular environment the growth and development of specific structure is controlled by various components such as growth factors/its receptors, signaling molecules, ECM proteins, adhesion molecules, morphogenic molecules, protein kinases, non-protein kinases, phosphatases, transcription factors, all contribute to the development.
Mol.biol; The cells to survive require a continuous supply of oxygen and nutrients ( "they need to eat"), which is carried by the blood. For this reason the blood vessels are the first organ that develops in the embryo and represent the most extensive network of our body. http://www.bioricrescita.it
This diagram just indicates how so many factors contribute to the functioning of the cell as a community and communicate with each other all the time including development and distress.

Typical animal cell structure containing 19 organneles; http://diseasespictures.com/

The plasma membrane shown contains receptors of different kinds. Examples of the action of transmembrane proteins; Transporters carry a molecule (such as glucose) from one side of the plasma membrane to the other. Receptors can bind an extracellular molecule (triangle), and this activates an intracellular process. Enzymes in the membrane can do the same thing they do in the cytoplasm of a cell: transform a molecule into another form. Anchor proteins can physically link intracellular structures with extracellular structures. © 2010 Nature Education All rights reserved; https://www.nature.com.

Transmembrane protein with several multipass domains with N terminal and c terminal on the opposite face of the membrane; https://themedicalbiochemistrypage.org/
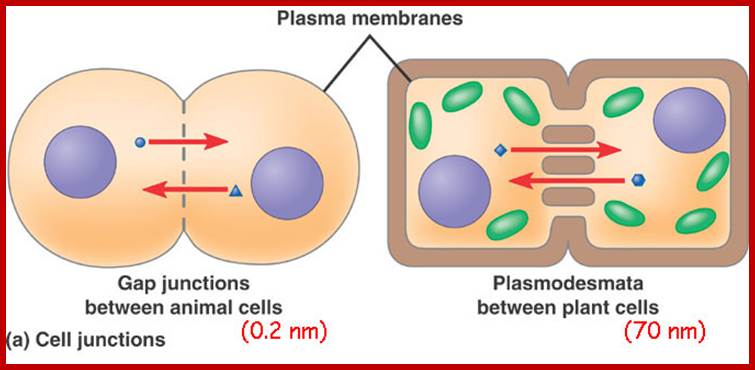
Cell communications: http://apbiobyanika.blogspot.com/

http://dauzbiotekhno.blogspot.com/
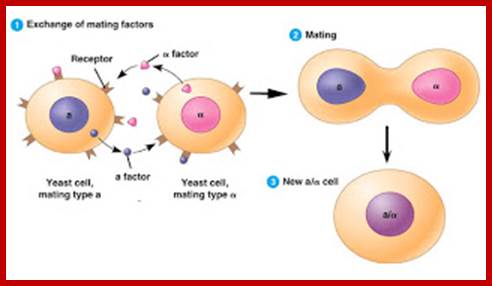
Cells of the yeast Saccharomyces cerevisiae use chemical signaling to identify cells of opposite mating type and to initiate the mating process. First cells of mating type A release a-factor, which binds to receptors on nearby cells of mating type B. Meanwhile, B cells release b-factor, which binds to specific receptors on A cells. Both these "factors" are small proteins of about 20 amino acids in length. Binding of these factors to the receptors induces changes in the cells that lead to their fusion, or mating. The resulting A/B cell combines in its nucleus all the genes from both A and B cells, (diploid).Communication between mating yeast cells: http://joosophie.blogspot.com/
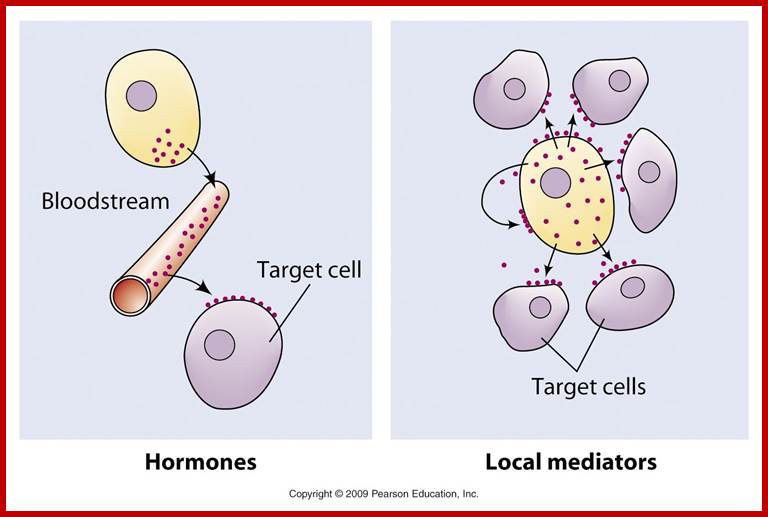
Endocrine systems and Hormones; http://www.biologyaspoetry.com/
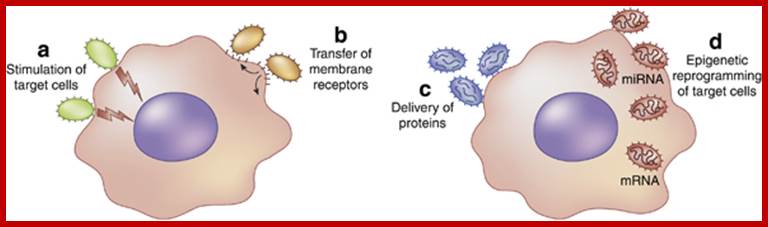
Exosomes/microvesicles as a mechanism of cell-to-cell communication; Schematic representation of mechanisms involved in micro vesicle (MV)-mediated cell-to-cell communication. (a) MVs may act as a ‘signaling complex’ through surface-expressed ligands that directly stimulate the target cells. (b) MVs may transfer receptors between cells. (c) MVs may deliver functional proteins or infectious particles to target cells. (d) MVs may transfer genetic information via mRNA, microRNA (miRNA), or transcription factors from one cell to another. Giovanni Camussi, Maria C Deregibus, Stefania Bruno, Vincenzo Cantaluppi and Luigi Biancone; http://www.nature.com/
Molecular Therapy: Microvesicle-mediated intercellular communication; Components of donor cells are incorporated into microvesicles (e.g., exosomes, shedding microvesicles, and apoptotic blebs) that contain proteins, e.g., signaling proteins, transcriptional regulators, reverse transcriptase, and transmembrane proteins; RNA (messenger RNA (mRNA), noncoding RNA (ncRNA), and microRNA (miRNA)); and DNA (genomic DNA (gDNA) and complementary DNA (cDNA)). Microvesicles may initiate signals through interaction between ligands on their surface and receptors on the recipient cell and/or have their contents taken up by recipient cells through endocytosis or fusion at the plasma membrane. (1) Transmembrane proteins can be transferred to the plasma membrane and trigger signaling. (2) Transcriptional regulators can be transferred into the nucleus and regulate promoter activity. (3) mRNAs/miRNAs can be transferred and influence the translational profile. (4) Donor cell–derived cDNAs (e.g., for c-Myc) can be delivered into the recipient cytoplasm (5) or generated from reverse-transcribed mRNAs. (6) Retrotransposon and other DNA elements from microvesicles may integrate into the recipient cell genome. These microvesicle delivery events have the potential to change the phenotype of recipient cells on a short- or long-term basis. (Reprinted with permission; http://www.nature.com/
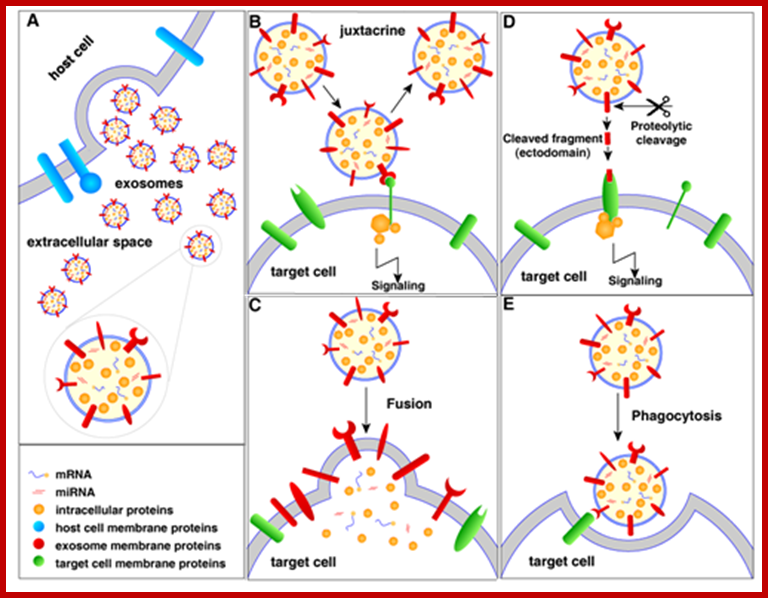
Extracellular vesicles (EVs) are membranous vesicles released by a variety of cells into the extracellular microenvironment. Based on the mode of biogenesis, EVs can be classified into three broad classes (i), ectosomes or shedding microvesicles (ii), exosomes and (iii), apoptotic bodies. Recent studies have ignited significant interest on EVs by elucidating their role in intercellular communication, pathogenesis, drug, vaccine and gene-vector delivery and as possible reservoirs of biomarkers. ; Community compendium of extracellular vesicle; Signaling of extracellular vesicles (exosomes as example); http://microvesicles.org/
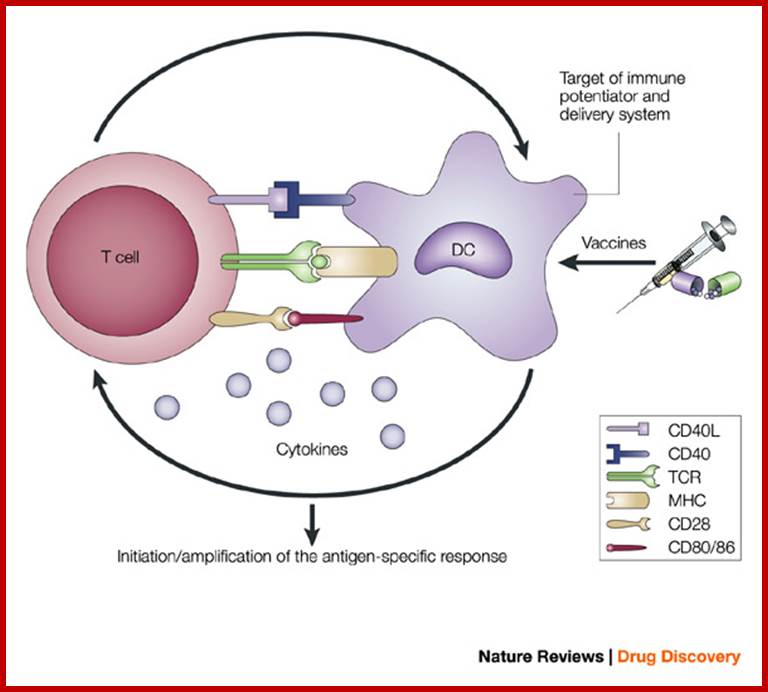
Antigen-presenting cells serve as the bridge between innate and antigen-specific responses.
The intimate interactions (cognate recognition) between antigen-presenting cells (APCs) and antigen-specific T cells initiate and amplify pathogen-specific responses. A dendritic cell (DC) is depicted here. The key interaction is driven by the recognition of antigenic peptide–major histocompatibility complex (MHC) dimers by T cells bearing T-cell receptors (TCRs) with high affinity for the complex. However, this signal alone is not sufficient for initiation and amplification of specific T-cell responses. Co-stimulatory signals (that is, CD28 recognition of CD80/CD86) and the production of pro-inflammatory cytokines, provide the 'infectious context' by which the full activation of antigen-specific T cells is achieved. The expression of co-stimulatory molecules and cytokines by APCs is tightly regulated and induced only when the APC encounters antigens associated with pathogen-associated molecular patterns. From the perspective of adjuvant design, the APC is a high-priority target. Delivery systems increase antigen uptake and presentation, and can also target immune potentiators more efficiently to APCs. Immune potentiators induce co-stimulatory signals and cytokine production, and the antigens select the highly specific T cells leading to the initiation and amplification of antigen-specific immunity. CD40L, CD40 ligand. Derek T. O'Hagan & Nicholas M. Valiante http://www.nature.com/
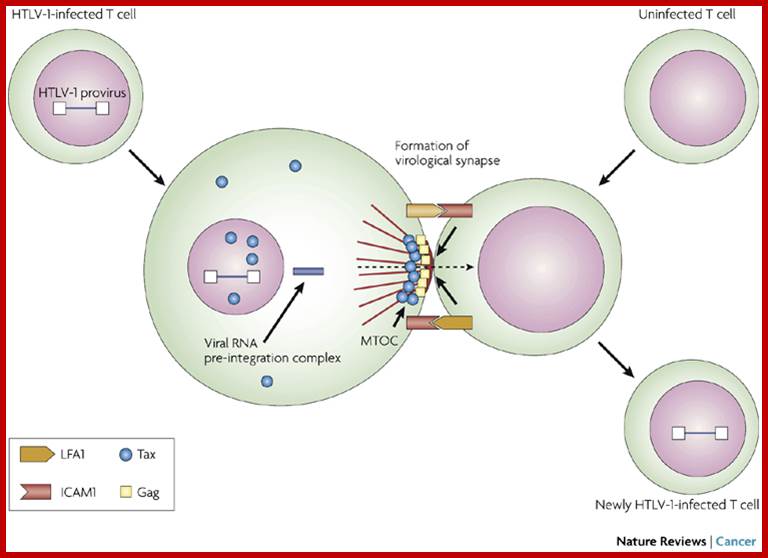
Cell–cell transmission of HTLV-1 occurs through a virological synapse. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation.
Unlike many other viruses, cell-free human T-cell leukemia virus type 1 (HTLV-1) virions are largely non-infectious. This function-associated antigen 1 (LFA1), and intercellular adhesion molecule 1 (ICAM1 figure illustrates the cell–cell contact required to create a virological synapse through which the viral genome is transmitted from one cell to another. The roles played by lymphocyte) in forming cell–cell contact are shown. Tax contributes to the formation of a microtubule organizing center (MTOC). Masao Matsuoka & Kuan-Teh Jeang http://www.nature.com/

http://dentistryandmedicine.blogspot.com/
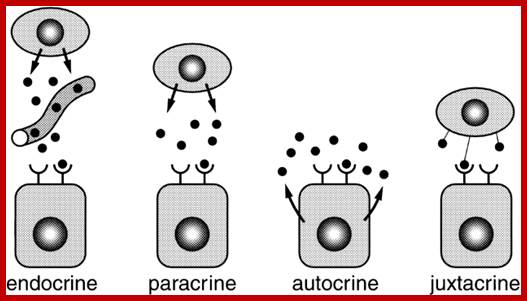
The secretion of chemical signalling molecules, the localization of receptors and the different signalling pathway types are shown. In case of autocrine signalling, the receptors are located on the secreting cell itself. In case of paracrine signalling, the receptors are located on neighbouring cells (tissue hormones). Endocrine glands secrete their products into the bloodstream; http://to.ttk.elte.hu
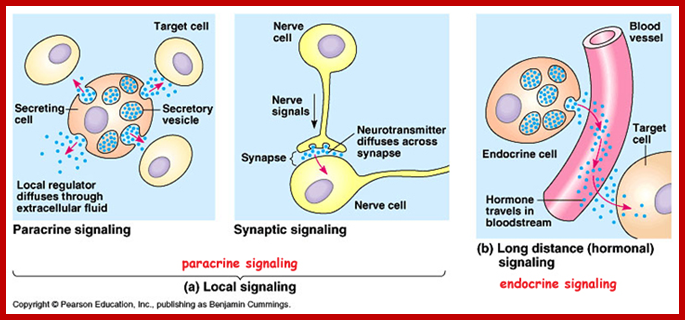
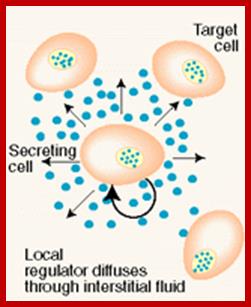
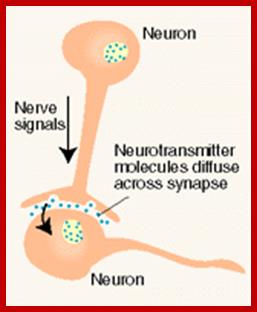
Paracrine, autocrine and synaptic signals: The local targeters; Paracrine, autocrine and synaptic are three types of local hormone signaling. In paracrine signaling, hormones are released into the fluid between cells (the interstitial fluid) and diffuse to nearby target cells. Hormones that influence secretions or other processes on the same cells that released them are said to be autocrine signalers. The more specialized synaptic signaling occurs between neurons (the nerve cells that make up the nervous system) and between neurons and muscle cells, allowing nerve cells to talk to each other and to muscles. http://www.oocities.org/
Important aspect of signals and receptors is that the receptors are specific to specific signals, with some variations. The number and the kind of receptors vary from cell type to cell type. The specific signal molecule binds to specific cell receptor and the receptor gets activated. The activated receptor sends the message across the cell and to the nucleus for genes to respond. The response is varied and specific, many a times the responses are multiple and run as cascades and amplify.
Bacterial signal transduction system:
Bacteria employ and use cell surface receptors. Bacteria too sense environmental factors, chemicals and cells respond to such signals, the signals can be in the form of chemicals, heat, light, water and toxins and others. Receptors for signals are located in the plasma membranes with external ligand binding exposed to periplasmic space and cytosolic transducing domain and a transmembrane anchoring domain. The binding of the signal, what is now can be called as ligand, induces receptor protein conformational change that is propagated across the membrane to cytosolic side of the protein. The conformation change induces and initiates signal transducing process. Most of the responses in bacteria such as osmoregulation, chemo taxis and sporulation and others are regulated by two component system. One of the components is sensor domain and the other is response domain. Phosphorylation of the inner regulator domain activates the cascade or activation at the cytosol. The signal is transferred from the input domain to transmitter domain, causes histidine p-lation, which in turn is passed on to the receiver domain; that leads to the output. The transmitter domain perse contains histidine kinase motifs. In general the receptor proteins work as dimers, in fact they become dimers with binding of ligands. The histidine kinase in turn phosphorylates aspartate at the cytosolic side of the receptor and it gets activated.
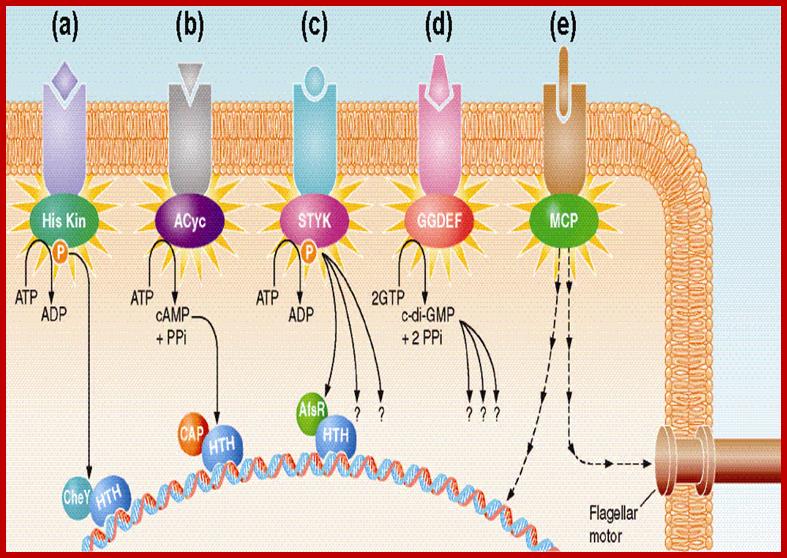
Comparative Genomics Approaches to Bacterial Signal Transduction;
A general view of bacterial transduction; majority of them shown act on Gene activation and one activate flagella movement. All signals act through activation of Kinase cascade. The complexity of bacterial signal transduction systems has long hindered their systematic analysis even in classical model microorganisms. The availability of complete genome sequences has changed that by allowing us to predict all (known) regulatory components in a given organism and see how much still remains to be learned about its bacterial signal transduction pathways. We employ comparative genome analysis to study common trends in environmental and intracellular signaling in various organisms, The key finding of this work was that microbial signaling systems are far more diverse than previously thought and include a network of diverse sensors that transmit signals to a variety of regulatory mechanisms. The total number of signal transduction proteins grows approximately as a square of genome size, whereas their distribution in various organisms suggests that different systems could cross-talk and compensate for each other. We have developed several metrics for genome comparisons, including signal protein family profiles, which reflect the abundance of each type of signal transduction proteins and could be used to trace their evolution in the course of adaptation to their specific ecological niches. Michael Y. Galperin; http://www2.eez.csic.es
Bacterial Signal Transduction: Two-Component Regulatory Systems (TCS) and Extra cytoplasmic Function (ECF) σ Factors:
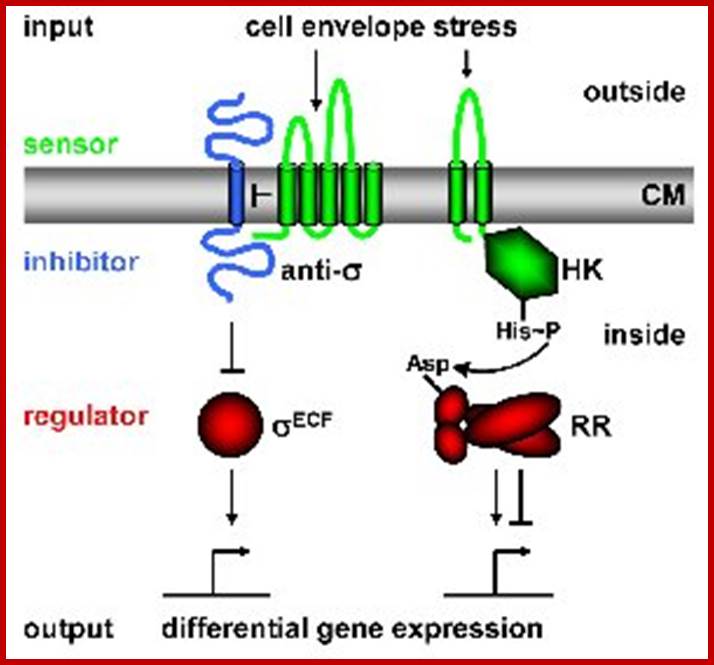 Bacterial Signal Transduction: Two-Component Regulatory Systems
(TCS) and Extra cytoplasmic Function (ECF) σ Factors; http://syntheticmicrobe.bio.lmu.de/
Bacterial Signal Transduction: Two-Component Regulatory Systems
(TCS) and Extra cytoplasmic Function (ECF) σ Factors; http://syntheticmicrobe.bio.lmu.de/
Stress response
In order to respond to changes in environmental parameters, cells must be able to transmit the information from the cell surface (site of induction) to the cytoplasm (site of cellular response). The two transmembrane signal-transducing mechanisms most common in bacteria are two-component systems and extra cytoplasmic function (ECF) σ factors.
Regulatory principles orchestrating bacterial stress responses: ECF σ factors (left) and TCS (right; HK, histidine kinase; RR, response regulator). Sensors are shown in green, inhibitors in blue, transcriptional regulators in red. Arrows indicate activation, T-shaped lines repression. CM = cytoplasmic membrane. The His-Asp phospho-transfer for TCS is indicated. Both systems consist of two proteins, a transmembrane sensor protein (histidine kinase and anti-s factor, respectively) that detects a specific stimulus (= input), and a cognate cytoplasmic effector protein (response regulator and σ factor) that mediates the cellular response, usually differential expression of target genes. The two systems differ in the way the two proteins communicate and their output. Two-component systems are activated and communicate through a series of phosphotransfer reactions, i.e. phosphorylation and dephosphorylation of both histidine kinase and response regulator (Parkinson 1993, Cell 73:857-71);.
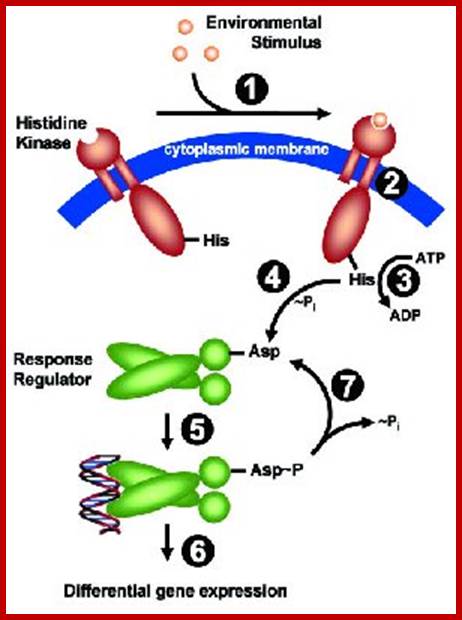
Two component signal transduction in bacteria: http://www.microbiologybytes.com/
Two component receptors:
Two-Component Systems (TCS) An archetypical system consists of (at least) two proteins, a sensor histidine kinase and a response regulator. Both proteins harbor two functional important domains: The histidine kinase detects a specific environmental stimulus through its (often periplasmic) sensor domain (input 1). This leads to a conformational change (2), resulting in ATP-dependent auto phosphorylation of a invariant His residue (3). His~P serves as the phosphate donor for the receiver domain of the cognate response regulator, resulting in phosphorylation of a conserved Asp residue and subsequently its dimerization (4). This ultimately leads to an activation of the effector domain of the response regulator, mediating the cellular response (= output 5), usually by mediating differential expression of specific target genes (= regulon 6). Set-back of the system to pre-stimulus state is achieved by de-phosphorylation, either by the intrinsic phosphatase activity exhibited by the transmitter domain of the sensor kinase, or by external phosphatases.
In contrast, ECF signal-transduction occurs through protein-protein interaction: In the absence of a signal the anti-s factor tightly binds the s factor, thereby keeping it in its inactive state. It is released in the presence of a stimulus and binds alternative promoter sequences upstream of its target genes (Helmann 2002, Adv Microb Physiol 46:47-110).
Two-component signal transduction systems: TCS is easily identifiable in genomic databases and they are present in more than 95% of the bacterial genomes sequenced to date as well as archaea and eukaryotic organisms such as plants and fungi. Cellular processes regulated by TCS are diverse, ranging from pathogenesis and cell development to metabolism and chemotaxis. Because of the widespread nature of TCS, researchers in many disciplines have reason to experimentally probe the molecular details of how a particular TCS functions in a system of interest.
Summary of useful methods for two-component system research:
Since the discovery of protein phosphorylation in bacterial nitrogen assimilation and chemotaxis more than 30 years ago, many biochemical techniques for the analysis of two-component signal transduction systems have been developed. Over the time experimental conditions to follow the flow of phosphate groups from histidine kinases to the cognate response regulators in vitro have been fine tuned. Several approaches were applied to circumvent the instability of the phosphorylated form of response regulator proteins to analyze the structures of their activated forms. Recently, a FRET (fluorescence resonance energy transfer) assay was developed to monitor interactions of chemo taxis proteins in vivo. The availability of bacterial genome sequence databases has facilitated the identification of two-component systems and enabled prediction of interacting kinase-response regulators pairs.
Two component systems in the spatial program of bacteria:
Despite being considered a relatively simple form of life, bacteria have revealed a high degree of structural organization, with the spatial destination of their components precisely regulated within the cell. Nevertheless, the primary signals that dictate differential distribution of cellular building blocks and physiological processes remain in most cases largely undisclosed. Signal transduction systems are no exception within this three-dimensional organization and two-component systems (TCS) involved in controlling cell division, differentiation, chemotaxis and virulence show specific and/or dynamic localization, engaging in the spatial program of the bacterial cell.

Signal transduction: Mechanism of ECF σ- dependent signal transduction: The three-stepped proteolytic cascade leading to anti-σ factor degradation and ECF σ factor activation is illustrated for σW. The stimulus is represented by the red arrow. Multipass or single pass membrane bound protein bound receptor. On the cytosolic surface one finds sigma factors associated with cytosolic domain of the membrane bound protein. Cytosolic proteins get degraded releasing sigma factor. Additional proteins are shown in yellow. The conserved alternative promoter sequence recognized by ECF σ factors is illustrated in the inset. The auto regulatory feedback loop (binding of the ECF σ factor to its own promoter) is indicated. http://syntheticmicrobe.bio.lmu.de/
The output of two-component systems is more flexible, allowing inhibition as well as induction of gene expression, while ECF s factors function as activators of expression only. The B. subtilis genome encodes 7 ECF σ factors and 35 two-component systems to sense changes in and communicate with the environment. Many of these systems are still of unknown function. www.lmu.de Prof. Dr. Thorsten Mascher.
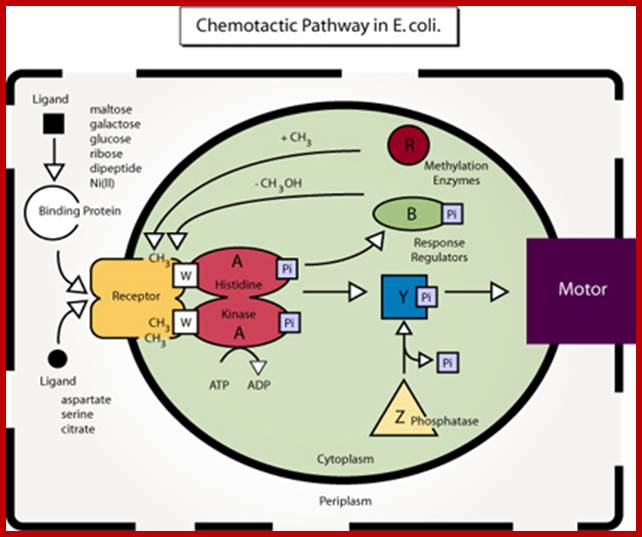
Diagram of the chemotactic pathway in E. coli. (Figure by MIT OCW: After figure 4 in Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. "The Two-component Signaling Pathway of Bacterial Chemotaxis: A Molecular View of Signal Transduction by Receptors, Kinases, and Adaptation Enzymes." In Annu Rev Cell Dev Biol. 13 (1997): 457-512.); http://2010.igem.org/
In bacteria, cell motility are a consequence of flagella rotation (in flagellated bacteria), which let them move in liquid mediums, and sometimes in solid mediums (agar plates). The name of these processes are swimming and swarming. However, there are others types of movements like: gliding movements (when cells have no cilium or flagella), twitching movements (intermittent, directionless and uncoordinated movement). Pseudomonas motility is due to the rotation of an only flagellum, H. salinarum motility is caused by a bunch of polarized flagella, but Escherichia coli, Salmonella, Bacillus subtillis have lots of flagella that are randomly spread out all over the membrane. Most of the time all flagella rotate in the same direction, provoking straight movements called run, but when the direction changes the bacteria begin to go round (tumbles). When ligand bound protein or ligand by itself binds to receptor activates Histidine kinase which is associated with the receptor gets activated by phosphorylation. This activates downstream events.

The mechanism of stimulus perception in bacterial two-component systems: Intramembrane-sensing histidine kinases; Stimulus perception: Schematic representation of the three different mechanisms of signal perception. (A) Periplasmic-sensing HK, (B) HK with sensing mechanisms linked to the transmembrane regions (signal perception can occur either with the membrane-spanning helices alone, or by combination of the transmembrane regions and short extracellular loops), (C) Cytoplasmic-sensing HK (either soluble or membrane-anchored proteins). The signal is represented by red arrows or red stars. The parts of the proteins involved in signal perception are highlighted in color.http://syntheticmicrobe.bio.lmu.de/
Rewiring Bacteria, Two Components signal Transduction at a Time; Michael A. Kohanski, James J. Collins,
To demonstrate the feasibility of rewiring two-component systems using their data, the authors focused on modifying the specificity of the osmolarity sensor protein from E. coli, called EnvZ, that normally targets the response regulator protein OmpR (Figure). To confirm their computational prediction that specificity is encoded by the phosphorylation subdomain, Skerker et al. first modified EnvZ by replacing its entire phosphorylation subdomain with either that of another E. coli sensor protein called RstB or that of a sensor protein called CC1181 from the bacteria Caulobacter crescentus. The authors found that the modified EnvZ sensor protein successfully phosphorylated the correct cognate response regulators of either RstB or CC1181 in vitro depending on the identity of the phosphorylation subdomain. Skerker et al. then further confirmed their prediction of the precise region in the phosphorylation subdomain that contains the residues required for sensor-regulator specificity. They replaced this region in the EnvZ phosphorylation subdomain with corresponding regions from five other E. coli two-component sensor kinases. These modified EnvZ proteins containing partial subdomain replacements also demonstrated the correct specificity in cognate response regulator phosphorylation.
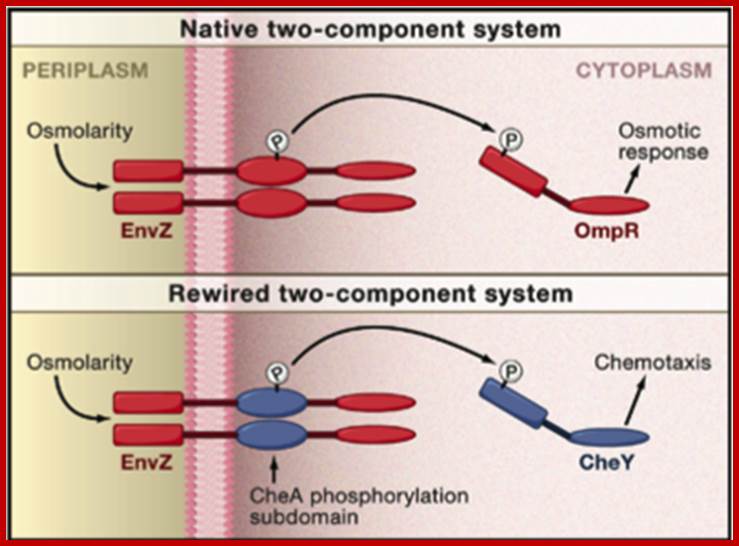
Rewiring a Bacterial Two-Component System(Top) Shown is the native EnvZ-OmpR two-component signaltransduction system (red) from the bacteria Escherichia coli for sensing and responding to osmolarity changes. (Bottom) A theoretical EnvZ sensor (red) rewired for chemotaxis pathway (CheA-CheY; blue) specificity, which still senses osmolarity changes but responds by initiating expression of chemotaxis genes. Jeffrey M. Skerker, Barrett S. Perchuk, Albert Siryaporn, Emma A. Lubin, Orr Ashenberg, Mark Goulian, Michael T. Laub
The two component system is well illustrated in E.coli, where it consists of two membranes with a periplasmic space between. Osmo regulation is controlled by two porins called OmpF and OmpC. OmpF is larger than OmpC and they are located in the outer membrane.
Osmoporin C: http://opm.phar.umich.edu/
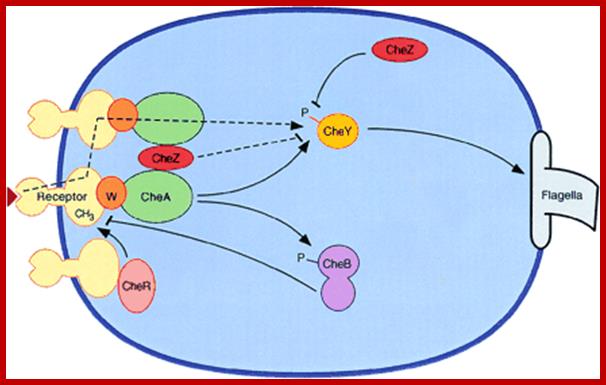
A nonlinear stimulus-response relation in bacterial chemotaxis; Schematic diagram of bacterial chemotaxis signal transduction. Transmembrane receptor proteins bind chemoeffectors and, through an adapter protein, CheW, control the activity of histidine protein kinase CheA. The cytoplasmic domains of the receptors are methylated by methyltransferase CheR and demethylated by methylesterase CheB. Attractant binding decreases kinase activity, while receptor methylation increases kinase activity. CheA provides phosphoryl groups to CheY and CheB, producing active forms of these proteins. Phosphorylated CheB demethylates receptors, providing a feedback loop that contributes to adaptation. The response regulator, phosphorylated CheY, binds to the flagellar motor, inducing clockwise flagellar rotation and a tumbling response. CheZ accelerates the dephosphorylation of CheY. The dashed lines indicate two postulated mechanisms for amplification of the excitation signal. Arrowheads and bars represent positive and negative regulation of phosphorylation, methylation, and clockwise flagellar rotation. http://www.pnas.org/
Chemotaxis via chemoreceptor:
When bacteria are subjected to higher osmomolarity, cells produce more OmpC resulting in small pores and they are expressed at the outer membrane. When bacteria is placed in low osmomolarity medium more OmpF are synthesized and localized in the outer membrane. The sensor protein EnvZ, for this kind of osmoregulation, is localized in the inner membrane. The N part of the sensor protein has a periplasmic domain and a transmembrane domain and the C-end has cytosolic domain. With osmomolarity the input domain undergoes conformational change and it is transduced to transmitter domain which gets auto phosphorylated at histidine residues. The phosphate is rapidly transferred to aspartate of the regulator OmpR. The N-part of the OmpR contains a DNA binding domain. When activated, the OmpR interacts with RNA polymerase and enhance the expression of OmpC and repress the expression OmpF. The osmoregulation started at external surface ends in gene regulation.
Signal Transduction in Eukaryotes:
Signal transducing system is evolved around a billion year ago and developed a very complex network of signal communication and expression of the same (believed to be so). The signal transductional network is pervasive and malfunction of one pathway has devastating effect. Though the animal system has a whole variety of signal transducing components, plants too have such signal transduction pathway and they may differ in structure and mechanism.
Signaling substances:
Signal molecules vary in composition, size and structure-such as hormones, growth factors, neurotransmitters, pheromones, smell, taste, sound, light, temperature, oxygen including death signals. Within the body certain hormones are released, they are grouped into Endocrines which targets tissues elsewhere. Paracrines after release act on nearby cells and autocrines act on their own leading to cell growth, proliferation and differentiation or cell death in developing organs. Autocrines act on their own cells. Impact of signals to just briefing the students is the effect on cells and their change in structure and function. Signal molecules come in various forms and structures. Some of them are free and some are bound to membranes. Epinephrine and EGF are surface ligands on proteolysis they are released to act on their respective receptors. Some signals are membrane bound and released on proteolysis e.g. Apoptosis inducing Fas ligands. Some of the extracellular matrix components act as ligands.
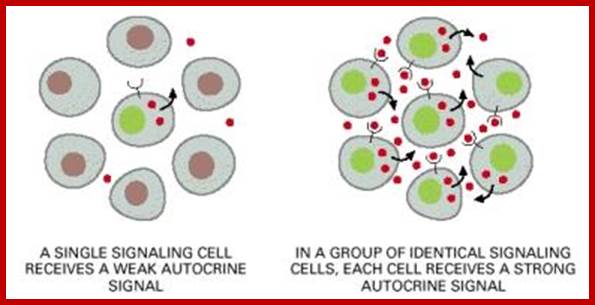
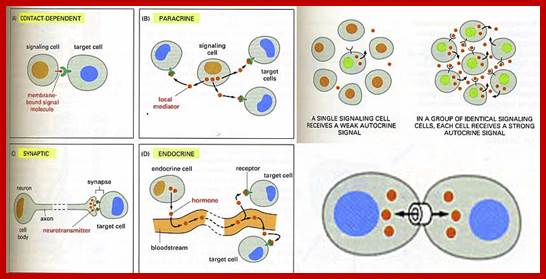
Autocrine, Paracrine and Endocrine mode of transmission. Signals and their effects on cells. Navrachana University; http://180.211.114.59:1111/;http://www.vcaspecialtyvets.com/
Signals and signal Molecules:
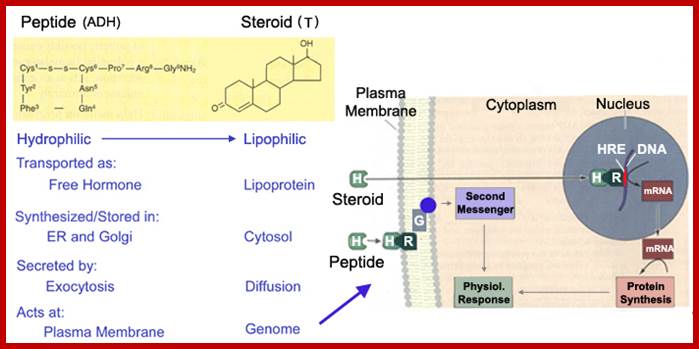
Signals can be light, temperature, wind, sound pathogens etc. Chemicals, based on the membrane solubility signal molecules can be classified, basically into two classes viz. water soluble (hydrophilic) and lipid soluble (lipophilic). Hydrophilic signals bind to membrane surface receptors and activate. Lipophilic molecules pass through lipid plasma membranes and enter cytoplasm and bind to receptors found in cytoplasm and activate them. Many steroid molecules binding to their respective cytosolic or nuclear receptors activate gene expression.
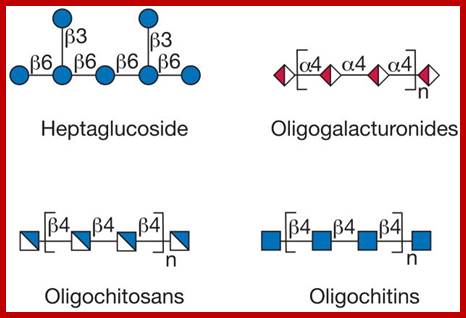

Glycans; Glycans on surface or inside the cells; https://www.intechopen.com
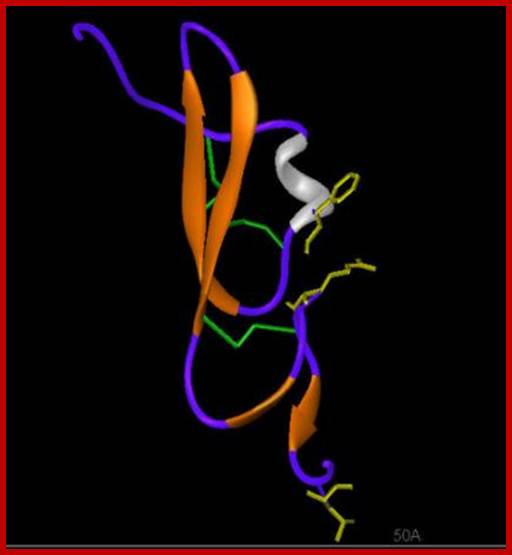
TGF alpha; http://www-nmr.cabm.rutgers.edu
(beta
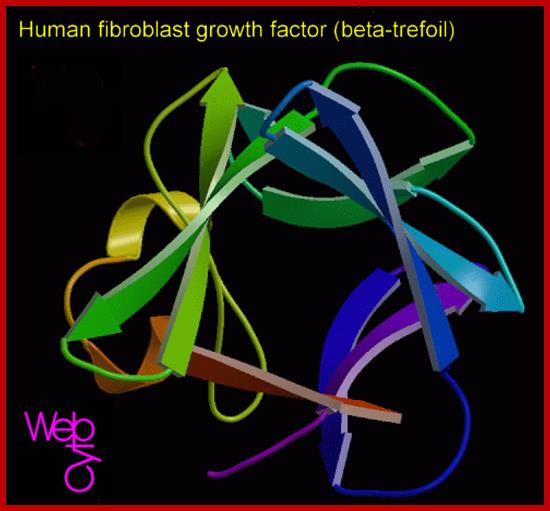
huFGFbeta-www.suggest-keywords.com
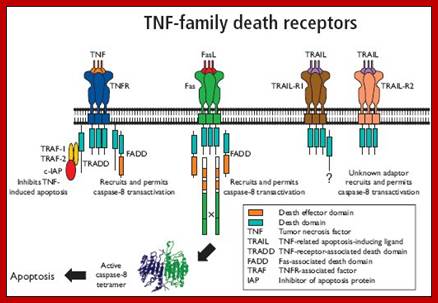
TNF alpha and Fas/Apo-1;https://www.bio-rad-antibodies.com
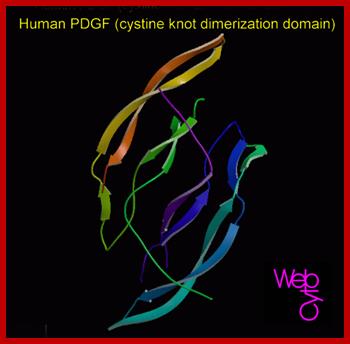
huPDGF; www.gtgwellness.com
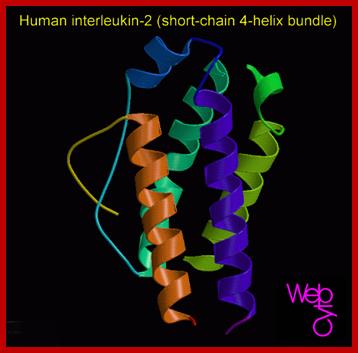

Hu Interleukin-2; http://diabetes.diabetesjournals.org
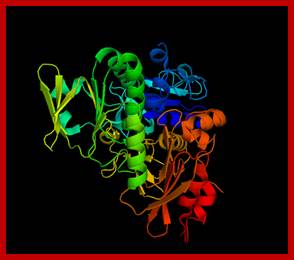

Apoptosis inducing factor (AIF); novel caspase-independent death effector released from mitochondria; Céline Candé et al, http://www.sciencedirect.com

Schematic representation of the intrinsic and
extrinsic apoptotic pathway. Apoptosis induction via the death receptor can
result in activation of the extrinsic and intrinsic pathways. Chemotherapy and
radiotherapy-induced apoptosis is executed via the intrinsic pathway. ![]() , activation;
, activation; ![]() , inhibition.
, inhibition.
Related Apoptosis-Inducing Ligand Pathway and Its Therapeutic Implications; Elisabeth G.E. de Vries, Jourik A. Gietema and Steven de Jong
Apoptosis-inducing factor (AIF) is a phylogenetically ancient mitochondrial intermembrane flavoprotein endowed with the unique capacity to induce caspase-independent peripheral chromatin condensation and large-scale DNA fragmentation when added to purified nuclei. In addition to its apoptogenic activity on nuclei, AIF can also participate in the regulation of apoptotic mitochondrial membrane permeabilization and exhibits an NADH oxidase activity. Under normal circumstances, AIF is secluded behind the outer mitochondrial membrane. However, upon apoptosis induction AIF translocates to the cytosol and the nucleus. Injection of anti-AIF antibodies or knockout of the AIF gene have demonstrated that AIF may be required for cell death occurring in response to some stimuli. In particular, inactivation of AIF renders embryonic stem cells resistant to cell death following growth factor withdrawal. Moreover, AIF is essential for programmed cell death during cavitation of embryoid bodies, the very first wave of (caspase-independent) cell death indispensable for mouse morphogenesis. We have recently found that AIF is neutralized by heat-shock protein (HSP) 70, in a reaction that appears to be independent of ATP or the ATP-binding domain (ABD) of HSP70 and thus differs from the previously described Apaf-1/HSP70 interaction (which requires ATP and the HSP70 ABD). Intriguingly, HSP70 lacking ABD (HSP70ΔABD) inhibits apoptosis induced by serum withdrawal, staurosporin, and menadione, three models of apoptosis which are also affected by micro-injection of anti-AIF antibody or genetic ablation of AIF. Altogether, these data suggest that AIF plays a role in the regulation of caspase-independent cell death.
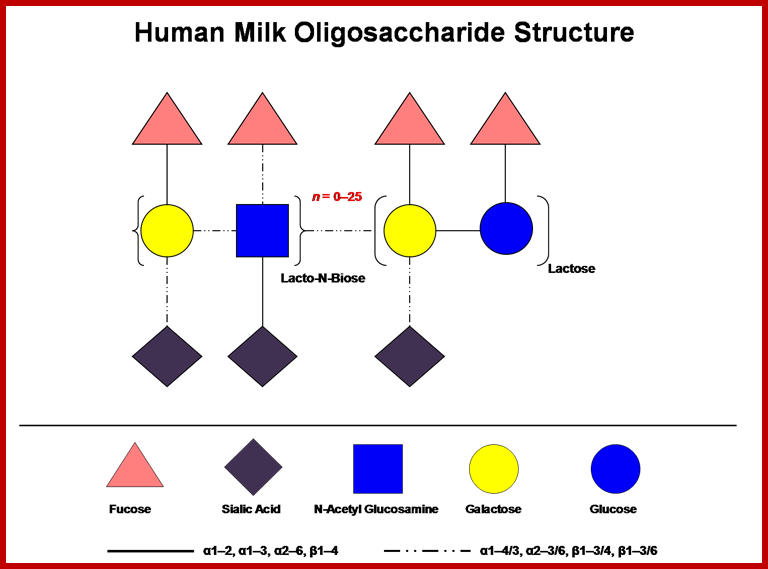
Human milk contains a high concentration of soluble oligosaccharides, which somewhat surprisingly exceeds the protein content (Coppa et al., 1993). The concentration of milk oligosaccharides vary by individual and declines over the course of lactation, but it is generally understood to be present at ≥ 4 g/L in milk with higher levels observed in colostrums (Asakuma et al., 2008; Chaturvedi et al., 2001). Milk oligosaccharides incorporate D-glucose, D-galactose, N-acetylglucosamine, L-fucose and sialic acid monosaccharides assembled in various configurations. Lactose (Galß1-4Glc) is predominately found at the reducing end and is elongated from a ß1-3 linkage (an additional ß 1-6 linkage is found in some branched forms) with several repeating N-acetyllactosamine units (Galß1-3/4GlcNAc). Further complexity is provided by terminal fucosylation via α1-2/3/4 linkages and/or α2-3/6 sialylation. Almost 200 distinct molecular species have been identified in pooled breast milk samples to date (Ninonuevo et al., 2006). When considering the immense combinatorial potential for these molecules, the identification of relatively few structures suggest that their biosynthesis within the mammary epithelia has been constrained by a functional imperative. Accordingly, HMO structures vary among maternal genotypes, with the particular complement of oligosaccharides present in mother’s milk dependant on Lewis and secretor status (Kunz et al., 2000). Further information on HMO structure and function is provided by recent reviews on the topic (Bode, 2006; German et al., 2008);http://milkgenomics.org/;Oligosaccharides; A cartoon depicting HMOs combinatorial potential
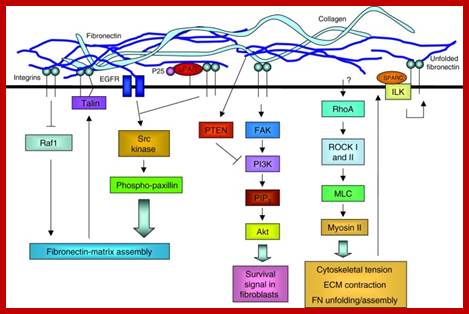
The extracellular matrix (ECM) regulates cell behavior by influencing cell proliferation, survival, shape, migration and differentiation. Far from being a static structure, the ECM is constantly undergoing remodeling – i.e. assembly and degradation – particularly during the normal processes of development, differentiation and wound repair. When misregulated, this can contribute to disease. ECM assembly is regulated by the 3D environment and the cellular tension that is transmitted through integrins. Degradation is controlled by complex proteolytic cascades, and misregulation of these results in ECM damage that is a common component of many diseases. Tissue engineering strives to replace damaged tissues with stem cells seeded on synthetic structures designed to mimic the ECM and thus restore the normal control of cell function. Stem cell self-renewal and differentiation is influenced by the 3D environment within the stem cell niche. For tissue-engineering strategies to be successful, the intimate dynamic relationship between cells and the ECM must be understood to ensure appropriate cell behavior. Extracellular matrix dynamics in development and regenerative medicine; William P. Daley, Sarah B. Peters, Melinda Larsen http://jcs.biologists.org
Lipophilic ligands:
The ligands are steroid components such as glucocorticoids, retinols, cholesterol derived compounds such as progesterone, androsterone, testosterone, estradiol and others.

Dexamethazone a corticoid; https://commons.wikimedia.org
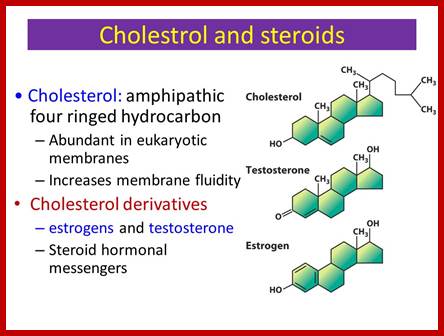
http://slideplayer.com/
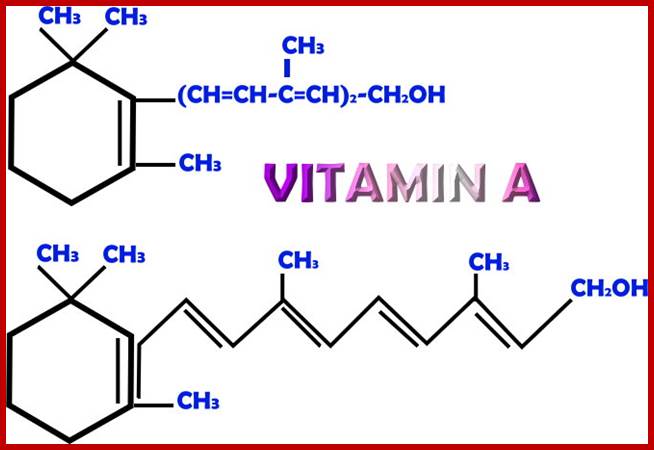

Vit A is important components of signal inducing substance. http://antioxidantsbiotech.yolasite.com https://phamix.com
Some cytosolic receptors after activation move into the nucleus, some are located in the nucleus and they get activated when ligands bind. The receptors most of them act as dimers and they contain domains like DNA binding, ligand binding and dimerization domain.
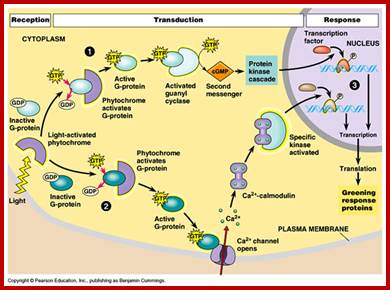
Light acts as signal transducing component; .DeepakYadav https://www.slideshare.net/
Receptors:
They are the structures that bind to signal molecules. Depending upon the type of signals such as Lipophilic and hydrophilic, one can discern the existence of two types of receptors. Most of the hydrophilic receptors are localized on the surface of the cells anchored to the membranes. The lipophilic receptors are found in cytosol or in the nucleus. The kind and the number of cell surface receptors varies from one cell type to the other, in extreme cases the number can be as large as 20,000 per cell. They act as the sentinel at the external cell surface.
Cell surface receptors:
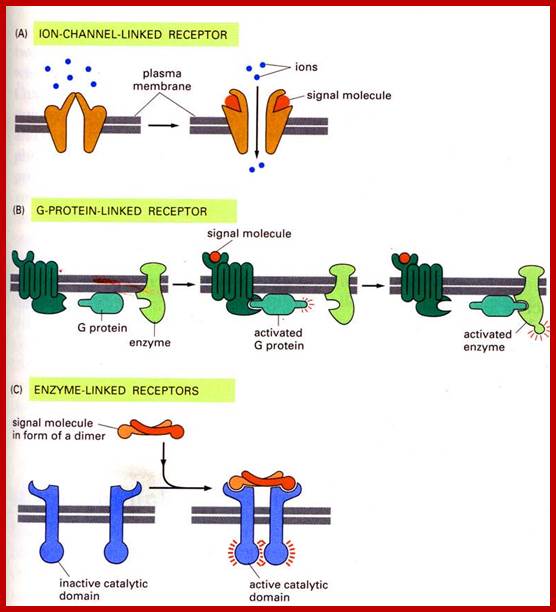
http://www.bio.miami.edu
Cell surface receptors can be ion channels, multipass membrane bound G-proteins or they can be cell surface bound receptors for specific ligands.
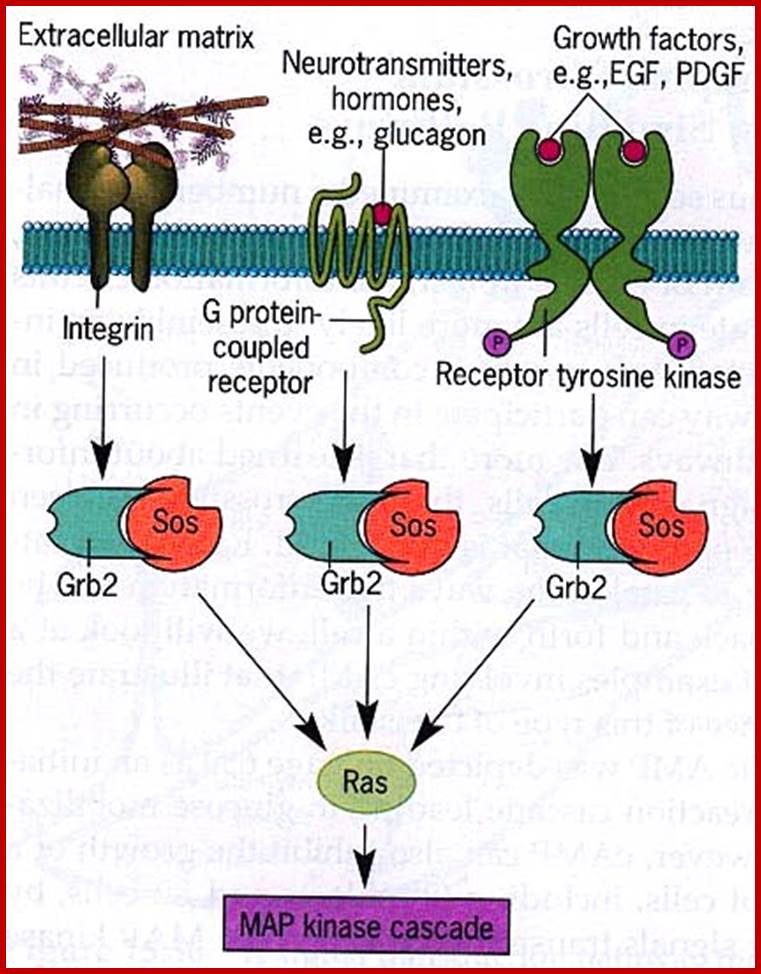
Different ligands bind to specific type of receptors; http://www.sivabio.50webs.com
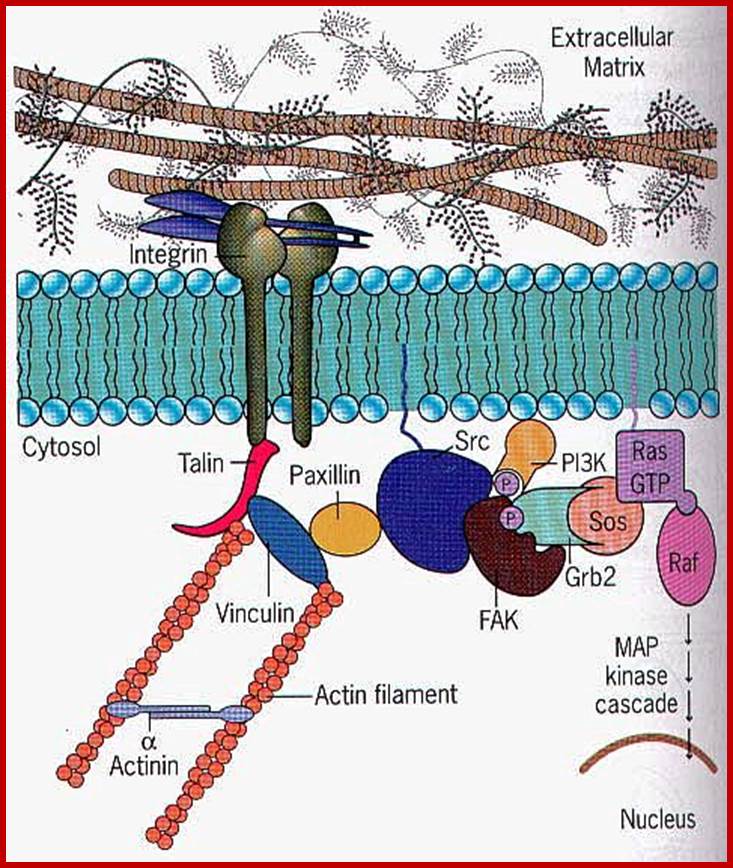
Cell signaling; Extra cellular matrix ligand activates cell surface receptor. http://www.sivabio.50webs.com/
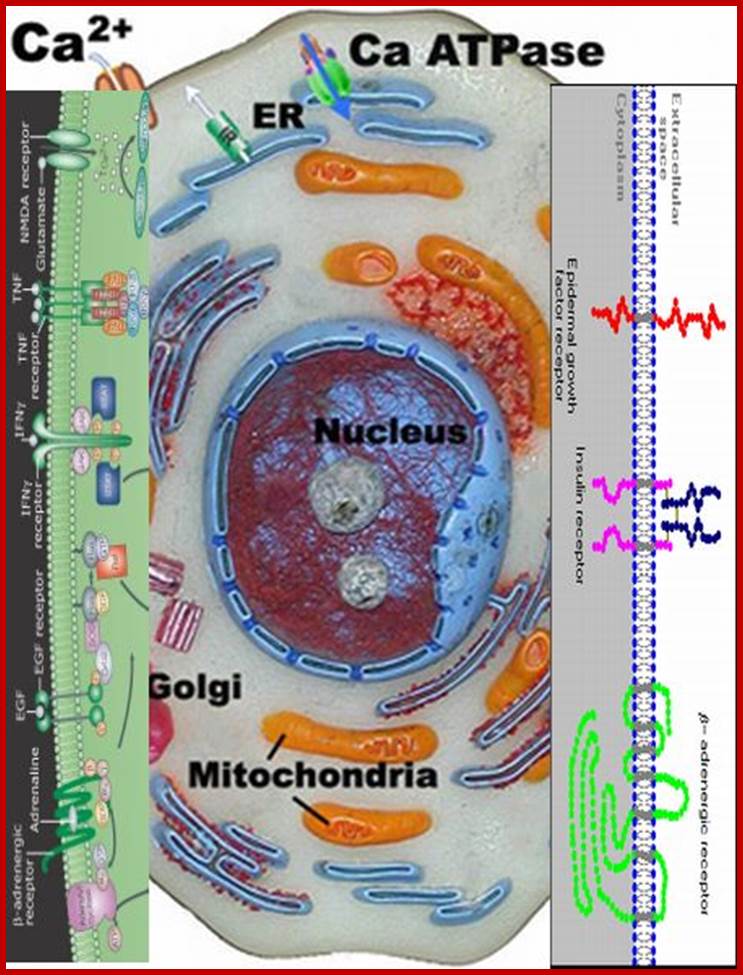
Very many receptors bound to cell membranes; they have extracellular surfaces and cytosolic surfaces; www.slideplayer.com

Surface receptor kinases- a plethora of them. ignaling molecules like proteins which are unable to enter into cells can interact with the cell surface receptors and execute its signaling process. Cell surface receptors are transmembrane proteins whose extracellular portion has the binding site for the signaling molecule and intracellular portion activates proteins in the cytosol that in different ways eventually regulate gene transcription in the nucleus; http://www.sivabio.50webs.com/
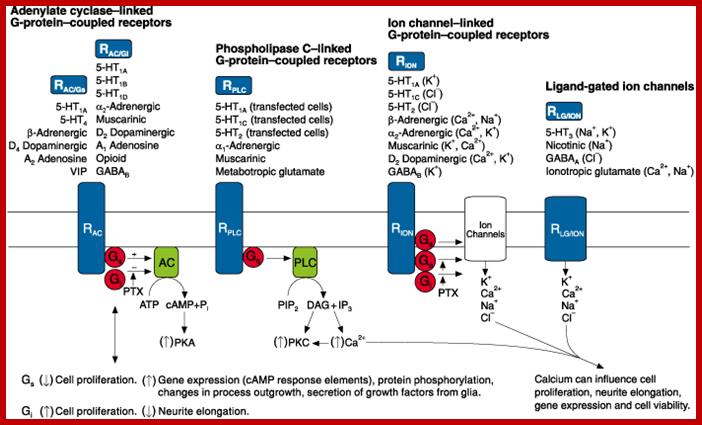
https://www.slideshare.net/
Different kinds of receptors such as cAMP cyclase linked G-protein coupled receptors, Phospholipase linked G-protein coupled receptors, Ion channel linked GPCR and Ligand gated ion channels
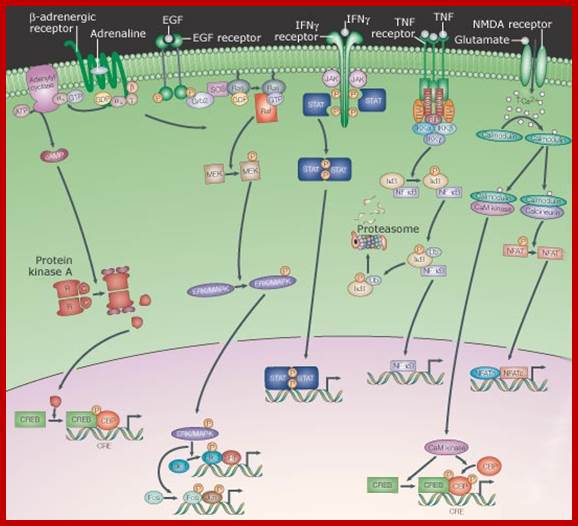
Signalling from the membrane to the nucleus, signalling from cell surface receptors through relay system; Internal signal transducers respond to External signals and get activated and then move into the nucleus where they activate specific genes. This group includes receptors that, after engaging their extracellular ligand, undergo a conformational change, which often induces them to dimerize or oligomerize. This results in the production or release of any of several second messengers - from ions to lipids - or the post-translational modification - from phosphorylation to proteolysis - of cytoplasmic proteins. A series of protein interactions and modifications then relays the signal - sometimes amplifying and diversifying it - to the nucleus, whereupon the activity of transcription factors is altered. As mentioned above, the pathways that are depicted have been necessarily simplified such that the arrows represent information flow. As such, the physical relationships between the components is largely ignored, although it is well established that localized complexes of components are important in the transduction of signals. http://opentextmining.org
|
Three subdivisions within this section can be made.
|
|
|
|
Acronyms: CaM kinase, calcium/calmodulin-dependent kinase; cAMP, cyclic AMP; CBP, CREB-binding protein; CREB, cAMP response element-binding protein; CRE, cAMP response element; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; Grb2, growth factor receptor-bound protein 2; IFN, interferon; IkB, inhibitor of nuclear factor kB; IKK, IkB kinase; JAK, Janus kinase; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; NFAT, nuclear factor of activated T-cells; NF-kB, nuclear factor kB; NMDA, N-methyl d-aspartate; RIP, receptor-interacting protein; SOS, son-of-sevenless; SRF, serum response factor; STAT, signal transducer and activator of transcription; TGFb, transforming growth factor b; TNF, tumour necrosis factor; TRADD, TNF receptor-associated via death domain; TRAF, TNF receptor; http://opentextmining.org |

Vascular endothelial growth factor (VEGF) is a key regulator of physiological angiogenesis during embryogenesis, skeletal growth and reproductive functions. VEGF has also been implicated in pathological angiogenesis associated with tumors, intraocular neovascular disorders and other conditions. The biological effects of VEGF are mediated by two receptor tyrosine kinases (RTKs), VEGFR-1 and VEGFR-2, which differ considerably in signaling properties. Non-signaling co-receptors also modulate VEGF RTK signaling. Currently, several VEGF inhibitors are undergoing clinical testing in several malignancies. VEGF inhibition is also being tested as a strategy for the prevention of angiogenesis, vascular leakage and visual loss in age-related macular degeneration.; VEGF responding cell surface receptors; http://www.nature.com/

Cell surface Interleukin receptors; The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design; Thomas A. Waldmann; http://www.nature.com
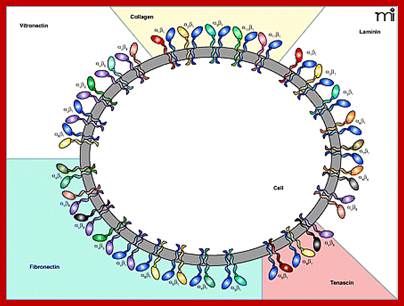

Matrix bound interacting integrin receptors; Senthil Arumugam, Patricia Bassereau; Membrane nanodomains; http://essays.biochemistry.org
There are non-receptor protein kinases such as Src kinases, they play very important role. Src non receptor kinase, the first cancer inducing gene discovered by Peyton Ross. Ras, Raf, Rap, MEK, ERK, MAPK, they are found in associated with inner surface of the membrane or free in cytoplasm.
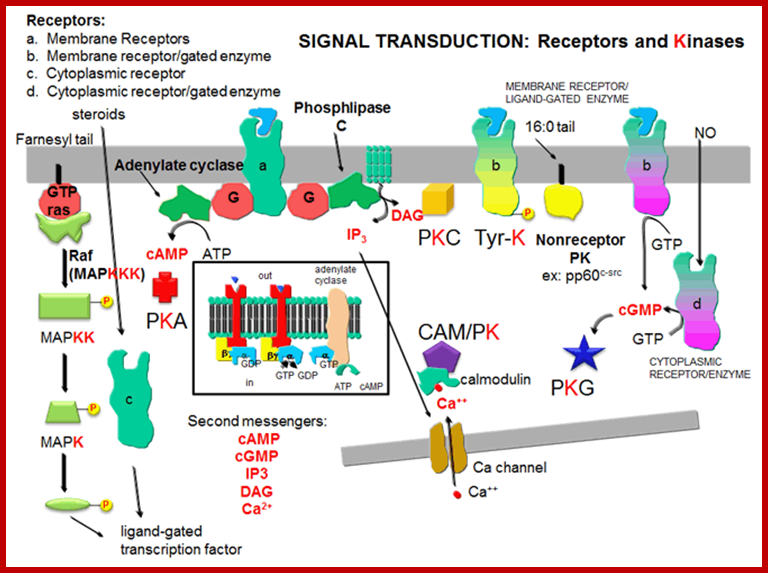
Signal transduction-Receptors and kinases; http://biowiki.ucdavis.edu/

Neurotransmission:Gating through G Linked Receptors; http://biowiki.ucdavis.edu/

Many proteins involved in signal transduction have SH2 domains. Some of these proteins also have catalytic domains with kinase activity. Others have phosphatase, transcription factor. or scaffolding domains, Ligand dependent protein kinases: http://biowiki.ucdavis.edu/

Receptor activated transportation; http://www.diabetes.diabetesjournals.org
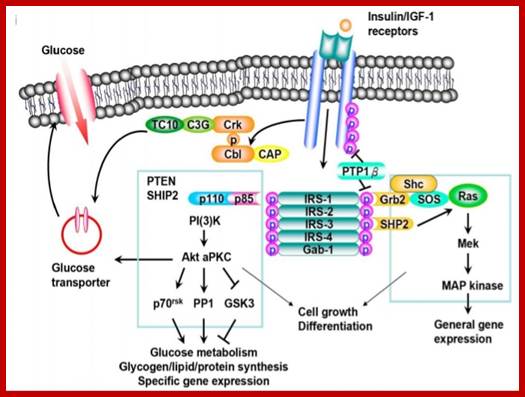
https://www.slideshare.net
Cell surface receptors; Types:
1. G-protein coupled Receptors (GPCR).
2. Receptor Tyrosine Kinases (RTKs).
3. Transforming Growth Factors (TGFb).
4. Hedge hog Receptors.
5. Wnt. Receptors.
6. Notch Receptors.
G-Protein Coupled Receptors:
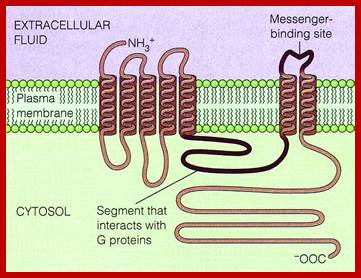
G-protein coupled receptors are largest class of receptors interact with cytosolic heteromeric G-protein. The G-PCR receptors are structurally more or less similar but functionally diverse. Their structure is similar to that of Bacteriorhodopsin (involved in bacterial photosynthesis) and also in vertebrate visual pigment of the eye and olfactory receptors of nose are surface receptors (a large class); consists of seven-transmembrane alpha helix domain with N-terminal exoplasmic and C-end cytoplasmic side. The G-protein binding site at helix loop between 6 and seven. Ligands bind to the exoplasmic surface of the receptor and activate G-protein at the cytoplasmic surface. The GPCR family includes receptors for a variety of hormones, neurotransmitters, light activated receptors and thousands of odorant receptors in nose.
Receptor activation is one of the primary steps that initiate the complex myocardial response to cardiac stress stimuli. G protein-coupled receptors play an important role in the initiation, regulation, and adaptation of cardiac hypertrophy. The activated receptors provoke intracellular signal-transduction pathways that carry out the hypertrophic response. Mitogen-activated protein kinases (MAPKs), protein kinase C (PKC), and calcineurin are examples of intermediate signaling pathways within the cytoplasm that directly modify transcription factors that promote alterations in the myocardial gene expression.
G protein-coupled receptors (GPCRs) are a large protein family that recognize extracellular molecules (i.e., ligands), activate intracellular activity, and ultimately initiate a cellular response. The GPCR is activated by an external signal in the form of a ligand or other signal mediator. The ligands particularly in the heart include hormones and neurotransmitters. When a ligand binds to the GPCR and is recognized, the GPCR makes a conformational (e.g., structural) change which mechanically activates the G protein.

G Protein-Coupled Receptor Activation and Initiation of the Intracellular Pathways; Myocardial GPCRs include adrenergic (comprised of α- and β-adrenergic receptor subtypes) and muscarinic receptors. These receptors are coupled to three principal classes of heterotrimeric GTP-binding proteins, Gs, Gαq/α11, and Gi,. These propogate the agonist- or antagonist-induced signal toward further internal effectors (i.e.,enzymes). All G proteins consist of Gα and Gβγ, subunits which upon activation dissociate and independently activate intracellular cascades.; http://pt851.wikidot.com
The G-proteins consists of three subunits such as alpha, beta and gamma. Mostly beta and gamma are bound together. The G-alpha is a GTPase switch protein. The G-protein gets activated with the binding of GTP.
Major classes of trimeric G Proteins:
|
G alpha |
Effector |
2nd messenger |
receptor |
|
Gs alpha |
Adenyl cyclase |
cAMP, up regulation |
b-adrenergic receptor,(epinephrine, glucagon) |
|
Gi alpha |
Adenyl cyclase, K+ channel protein |
cAMP change membrane potential-down |
Alpha1 Adrenergic receptor,mascrinic acetyl choline receptor |
|
G-alfa-alpha |
Adenyl cyclase |
cAMP, up |
Odorant receptor in nose |
|
G-q alpha |
Phospholipase C |
IP3, DAG receptors bind to G-protein complex -up |
Alpha2-adrenergi receptor |
|
G-o alpha |
Phospholipase C, |
IP3, DAG up |
Acetyl choline in, epinephrine cells |
|
G gamma alpha |
cGMP phosphodiesterase |
cGMP-down |
Rhodopsin in rod cells-light receptor |
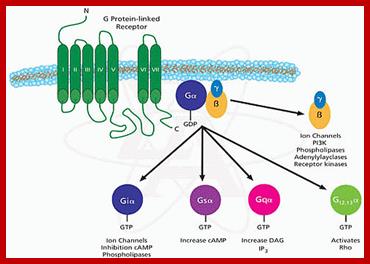
G-Protein linked multipass membrane receptor activates cytosolic G-protein components. http://repositorio.ul.pt/http://repositorio.ul.pt/ http://repositorio.ul.pt/
Human genome codes for 27 different G alpha, 5Gbeta and 13Ggamma subunits. Binding of the ligand to the receptor makes the receptor activated which in turn binds to G-complex and G-alpha becomes active with the binding of GTP, and G-alpha-GTP dissociates from the beta-gamma dimers complex. Both are anchored to the cytosolic surface of the plasma membrane. The G-alpha is monomer with 42-45kDa. Activated G-alpha GTP interacts with adenyl cyclase, which cyclizes ATP into cAMP which act as second messenger. The activated G-GTP is short lived. Activation makes the G-alpha-GTP to G-alpha GDP, which is inactive. And now it associates with G-beta and gamma as an inactive trimeric complex.
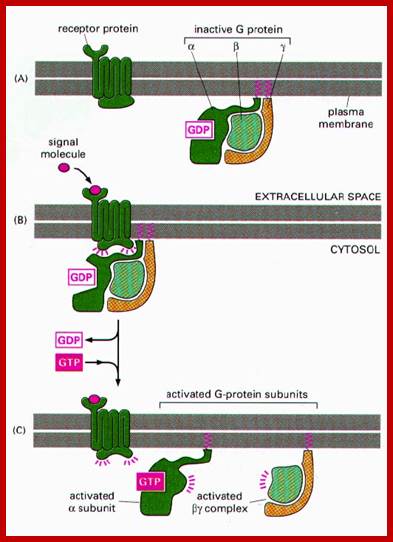
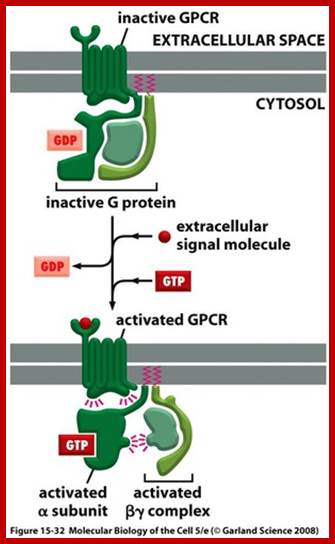
http://quizlet.com/
Activated G-protein activates adenyl cyclase resulting in the synthesis of cAMP, binds to an inhibitor that is bound to PK-A and releases PK-A from inhibition. The activated PK-A enters the nucleus and activates CREBp, which binds to enhancer region of specific genes and activates transcription.

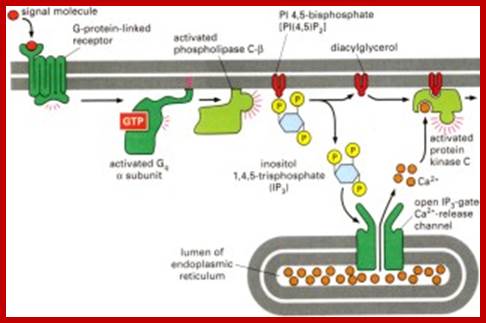
.A typical signaling pathway after activation of a GPCR receptor: Upon binding, the receptor activates several G-proteins and these activate other second messenger. In this way, the original signal is branched, amplified and translated into a cellular reaction (taken from Alberts et al., The molecular Biology of the Cell, 2002).Activated receptor transducing the effect that activates nuclear genes. Martin.Hegner/ http://www.tcd.ie/Physics/ Membrane_protein_background.html
The GPCR receptor is exemplified by epinephrine adrenergic receptor. Such receptors are found on hepatic cells, adipose tissue. Binding of epinephrine increases the heart beat (say contraction); activation of beta adrenergic receptors in smooth cells makes it relaxed. Adrenergic receptor found in on smooth muscle cells lining blood vessels in intestinal tract, skin and kidneys, on binding to the said hormone causes arteries to contract cutting off the supply of blood to peripheral organs. Epinephrine has diverse effect on diverse receptor structures, but in all these the effect is to provide energy supply in the form of substrates. Epinephrine receptors are GPCR receptors, but vary depending upon different types of Beta1 and beta2 type of adrenergic receptors coupled with G-proteins, that activates adenyl cyclase releasing cAMP, thus epinephrine increases the levels of cAMP. On the contrary alpha1 and alpha2 adrenergic receptors are coupled to with different forms of G-proteins. Alpha1 is coupled GI that inhibits adenyl cyclase. In contrast Gq coupled to alpha2 adrenergic receptor activates different effector enzymes that generates different second messengers.
The G trimeric proteins are versatile, and each kind interacts with different forms of receptor proteins. In liver, glucagon and epinephrine bind to different receptors and interact and activate the same Gs and trigger the adenyl cyclase and trigger the same response. In adipose tissue stimulation by epinephrine and glucagon or ACTH activates adenyl cyclase, whereas prostaglandin PGE1 or adenosine inhibits the enzyme.
Cholera toxin (Vibrio cholera) binds to G-GTP protein and prevents dissociation of GTP, thus causes expulsion of water and electrolytes into the intestine. Bordello toxin prevents the release of GDP thus locks up G1alpha, causes increased cAMP and promotes loss of fluids and electrolytes and mucus from the epithelial airways.
Adrenergic receptor proteins 42-45Kda consists of seven membrane pass proteins with N-terminal exoplasmic and C-terminal towards cytosol. The third C-loop is long and variable. It is to this C5 loop various components of G-protein complexes bind. The C2 loop also contributes to interaction with G-complexes. All four transmembrane helices contribute to ligand binding. G-alpha consists of six groups of alpha helices with two cytosolic globular domains, one between the first and second block and the second at C-end. Alpha3 and beta5 of this contacts adenyl cyclase when G-protein is activated (i.e. G-GTP). Ex, glucagon and epinephrine bind to different receptors but both activate the same Gs which activate adenyl cyclase, which triggers massive metabolic response.
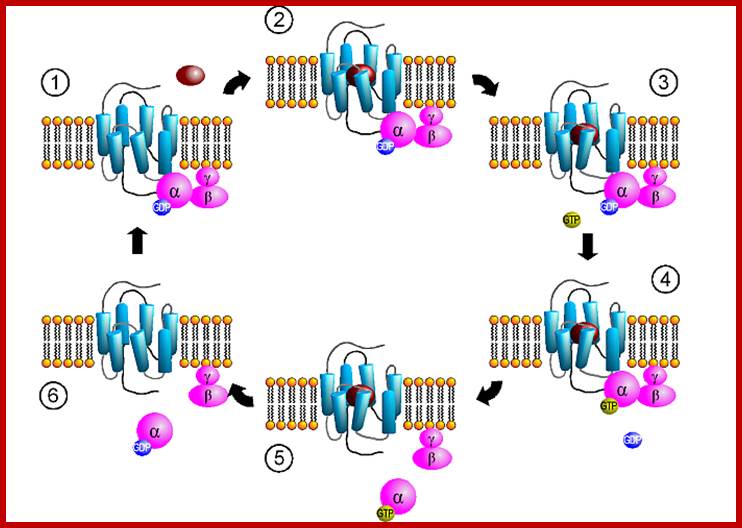
These G-proteins go through activation and inactivation cycle with the binding of ligands and dissociation of ligands. http://www.mun.ca/
www.medical-institution.com
Cell surfaces are always studded with different receptors, and the number of all receptors together can vary from 200 to 20000; it depends upon the kind of cell type and the need. The numbers of receptor at any given time go up or go down. Both RTKs and GPCR work simultaneously leading to concerted action and response.
Heterotrimeric G protein in vertebrate vision:
The G-protein in vertebrates activates cGMP phosphodiesterase during vision activities. Human eye consists of two types of photoreceptor cells, Rods and Cones. Rod is responsible for monochromatic vision in dim light, but cones are involved color vision in bright light. Rod cells are specialized in containing elongated stacks of densely packed membrane sacs called discs located at cellular tips, typical of granal stacks in chloroplasts. The stacked membranes contain photosensitive protein pigments called Rhodopsin. It is members of seven pass spanning transmembrane receptor family of proteins. Rhodopsin consists of a protein ‘Opsin” covalently linked light absorbing molecule 11-cis-retinal. When id absorbs single photon (400-600nm) it gets isomerized to all trans-retinal, this leads to conversion of the protein into meta rhodopsin (activated form). The activated form reduces the level of cGMP (which is synthesized by guanylate cyclase). In dark, cGMP builds up in the rod cells. cGMP gated channels keep opened for Na to be transported. This happens when the light is off; this leads to the depolarization of the membrane potential. When opsin becomes activated by light, it binds to heterotrimeric G-protein called ‘Transducin”. This causes G alpha subunit exchange GDP to GTP and dissociates from the trimeric complex. The G-alpha-GTP of transducin activates 3’5-cGMP to 5’-GMP, thus light causes decrease in the concentration of cGMP, thus it causes gated Na+ channels to close. The level of cGMP is controlled by guanylyl cyclase and cGMP Phosphodiesterase.
Cytosolic Receptors:
Most of the lipid soluble signals penetrate the plasma membrane and enter the cytosol where many kinds of receptors are found depending upon the cell type and the organism.
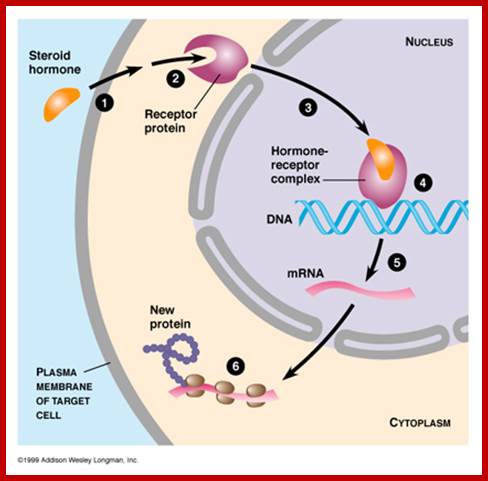
http://virtuallaboratory.colorado.edu/
Steroids, (testosterone, estrogens, cortisols) Type I ligands bind to inactive cytosolic receptors for they are bound to heat shock proteins. Binding of the hormones activate them by dissociating heat shock proteins from the receptors. These activated receptors act as TFs, move into the nucleus and bind to their specific hormone response elements (HRE), which are mostly located in the enhancer region of the gene promoter/regulatory regions. Vitamin D, thyroid hormones and retinoids-Type II ligands move into the nucleus, where they bind to their nuclear receptors and dimerize and bind to their respective response elements of the genes.

The neuronal synapse and the gap junction are one extreme in a signaling spectrum. At the other end are the endocrine or systemic signaling systems. Between these two extremes are juxtacrine or neighboring and paracrine or local signaling systems. Endocrine signals are secreted by cells directly into blood stream, travel throughout the body and affect many different cell types; these signalling molecules fall into two generic types, hydrophilic and hydrophobic moleculaes. Hydrophilic signaling molecules, or ligands, act by binding to receptor proteins on the surface of target cells. Hydrophobic ligands are transported through the blood stream attached to a hydrophilic carrier; http://virtuallaboratory.colorado.edu/
Receptors, most of them are all proteins, found located in cytoplasm in bound form or in free state. Many of them are located in the nucleus bound to their promoter regions, but in an inactive state. These become active only when their ligands, most of them are lipophilic, move into the cell and bind to their respective receptor and activate them.
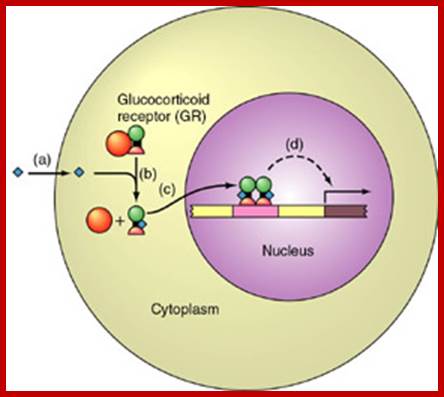
Cytosolic receptor protein; The Glucocorticoid receptors: Circulation of the GRa between the cytoplasm and the nucleus, and its transactivating or transrepressive activities. Upon binding to ligand, GRalpha dissociates from HSPs and translocate into the nucleus, where it regulates the transcriptional activity of glucocorticoid-responsive genes positively and negatively either by binding to GREs located in the promoter region of target genes or by physically interacting with other transcription factors. After completion of changing the transcriptional activity of glucocorticoid-responsive genes, the receptor is exported into the cytoplasm and is incorporated into the complex with HSPs. Abbreviations: GR: glucocorticoid receptor; GRE: glucocorticoid responsive element; HSPs: heat shock proteins; TF: transcription factor; TFREs: transcription factor responsive elements.; Tomoshige Kino; http://brainimmune.com/
Tomoshige Kino; http://brainimmune.com/
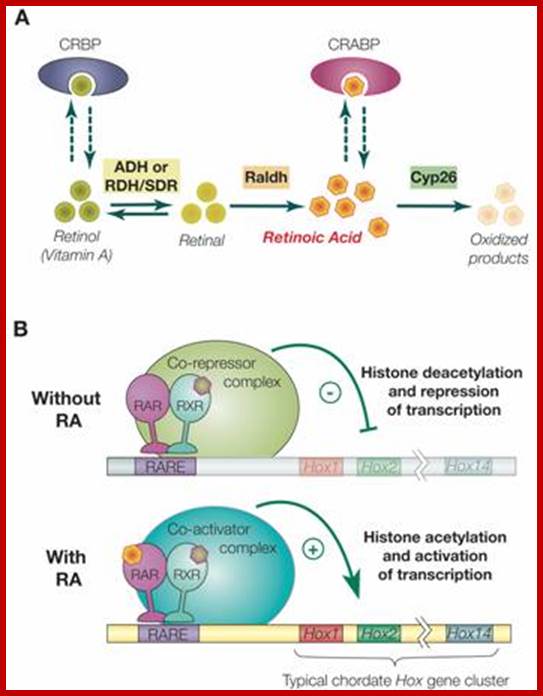
Retinoic acid induced gene activation of Hox genes. Ferdinand Marlétaz1, Linda Z. Holland et al; http://www.ijbs.com/Int J Biol Sci 2006;
Synthesis, degradation and mode of action of retinoic acid (RA): A) The metabolic pathway for synthesis and degradation of endogenous RA is shown. RA is synthesized by oxidation of retinal by retinaldehyde dehydrogenases (RALDHs). In a reversible reaction, retinal is synthesized from retinol (vitamin A) by either aldehyde dehydrogenases (ADHs) or short-chain dehydrogenase/reductase (RDHs/SDRs). Cellular retinol binding proteins (CRBPs) can bind retinol, whereas cellular retinoic acid binding proteins (CRABPs) can bind RA. Finally, endogenous RA is degraded by CYP26 enzymes. B) The RAR/RXR heterodimer mediates the effects of RA. In the absence of ligand (RA), the RAR/RXR heterodimer is bound to DNA and co-repressors. This complex induces transcriptional repression through histone deacetylation. Binding of the ligand (RA) induces conformational changes and the binding of co-activators leading to histone acetylation and activation of transcription.
RA signaling is mediated by RA binding to retinoic acid receptors (RARs), which form heterodimers with retinoid X receptors (RXRs). This complex in turn binds to retinoic acid response elements (RAREs) in the regulatory regions of target genes
In general, RAR and RXR proteins share a common organization of functional domains: an amino terminal A/B region containing a transcriptional activation domain (AF-1), a centrally located C region corresponding to the DNA binding domain (DBD) plus a weak dimerization domain and the E region, which includes the ligand binding domain (LBD), a strong dimerization interface and a surface allowing binding of transcriptional co-regulators. In the absence of ligand, the RAR/RXR heterodimer is constitutively bound to DNA on RAREs and associated with co-repressor complexes that induce transcriptional silencing by deacetylating histones associated with the target sequences thus increasing chromatin condensation. These co-repressors include the related proteins SMRT and NcoR. Binding of RA to the RAR ligand binding pocket induces a conformational change of the LBD that creates a surface allowing the association of co-activators and the release of co-repressors. The co-activators (e.g. TIF2 and SRC-1 of the p160 co-activator family) subsequently mediate histone acetylation resulting in decondensation of the chromatin and activation of target gene expression; Ferdinand Marlétaz1, Linda Z. Holland2, Vincent Laudet1, Michael Schubert
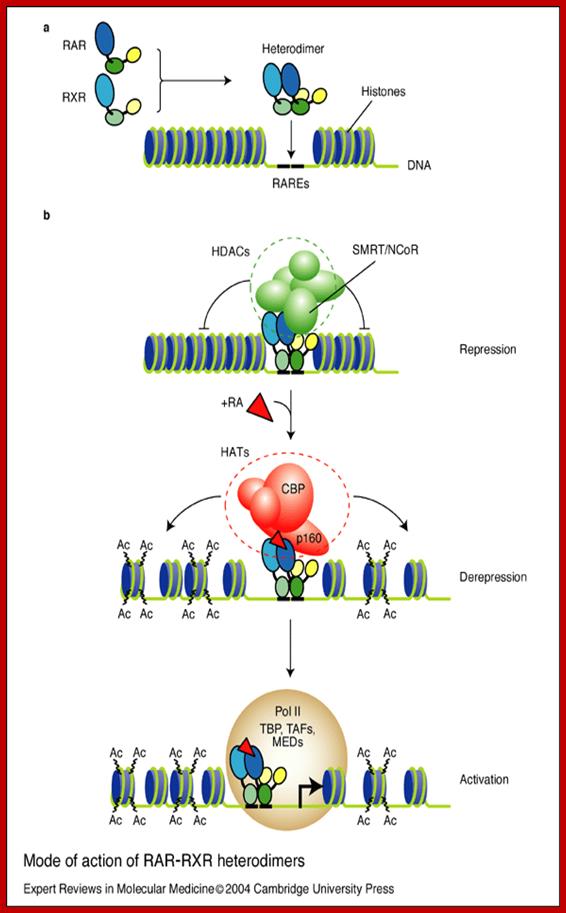
Mode of action of RAR–RXR heterodimers; Nicole Clarke et al; http://journals.cambridge.org/
Mode of action of RAR–RXR heterodimers: (a) Retinoic acid receptors (RARs) and retinoid X receptors (RXRs) form heterodimers that bind within the regulatory region of target genes through the retinoic acid response elements (RAREs). These cognate binding sites on DNA correspond to a 5 bp-spaced direct repeat (generally referred to as DR5) of polymorphic arrangements of the canonical motif 5'-PuG(G/T)TCA (shown as heavy black lines). (b) In the absence of ligand (e.g. RA), RAR–RXR heterodimers (apo-heterodimers) are believed to be bound to RAREs of target genes together with transcriptional co-repressors (N-CoR or SMRT), which then recruit histone deacetylases (HDACs). This is thought to account for a gene-silencing effect of apo-heterodimers through chromatin condensation (repression). Binding of ligand induces the release of the HDAC complex and results in the recruitment of histone acetyltransferase (HAT) co-activators, such as CBP and p160. The subsequent chromatin decondensation (derepression) due to histone acetylation (‘Ac’) is thought to be necessary, but not sufficient, for target gene activation. As the last step, the RNA polymerase II holoenzyme, together with the TATA-binding protein (TBP) and TBP-associated factors (TAFs), and mediator complexes (MEDs), are recruited, which increases the frequency of transcription initiation. Although the temporal order of factor recruitment is being determined with increasing precision, the composition of the complex and the dynamics and specificities of the events are largely elusive. Abbreviations: CBP, CREB-binding protein; NCoR, nuclear receptors corepressors; p160, co-activator (e.g. TIF2/RAC3/SRC-1); SMRT, silencing mediator for retinoid and thyroid hormone receptor. Nicole Clarke, Pierre Germain, Lucia Altucci and Hinrich Gronemeyer; http://journals.cambridge.org/
Note: This write-up is only for my students; it has been referred to and extracted from thousands of authors, whom I and my students are grateful. It is exhilarating to read some author’s write-ups directly, for each have their own way of unique expression and they have illustrated their concepts in great format. My writing is simple and direct. I am not an expert in all fields, but as I am teaching these subjects for nearly 50 years, I have collected lot of material as my teaching notes and the same is placed it for students of India and abroad.
